Figure 7. Rpp30 mutation leads to tRNA processing and piRNA transcription defects.
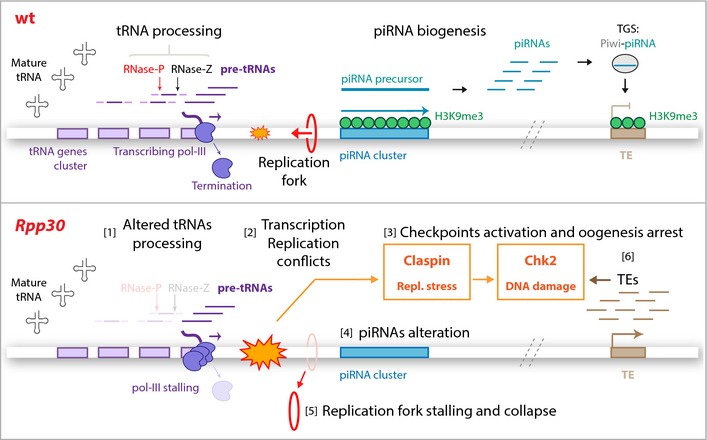
Top: In wild‐type ovaries, tRNA genes (violet boxes) are transcribed by pol III. tRNAs are then processed by RNase P and Z ribozymes that cleave tRNAs precursors (pre‐tRNAs, dark violet). tRNA gene clusters can be localized near piRNA clusters (turquoise), which are characterized by H3K9me3 epigenetic mark (green) required for piRNA transcription. piRNAs arising from piRNA precursors guide Piwi proteins to transcriptionally silence transposable element expression (TGS), protecting genome integrity. Bottom: In Rpp30 ovaries, tRNA processing defects [1] lead to transcription–replication conflicts (orange) [2]. These conflicts activate both replication stress and DNA damage checkpoint proteins, Claspin and Chk2 [3], which induce piRNA transcription defects [4] and replication fork stalling and PCNA collapse [5]. Downregulation of piRNAs results in TEs overexpression [6], which also activate DNA damage checkpoint [3].
