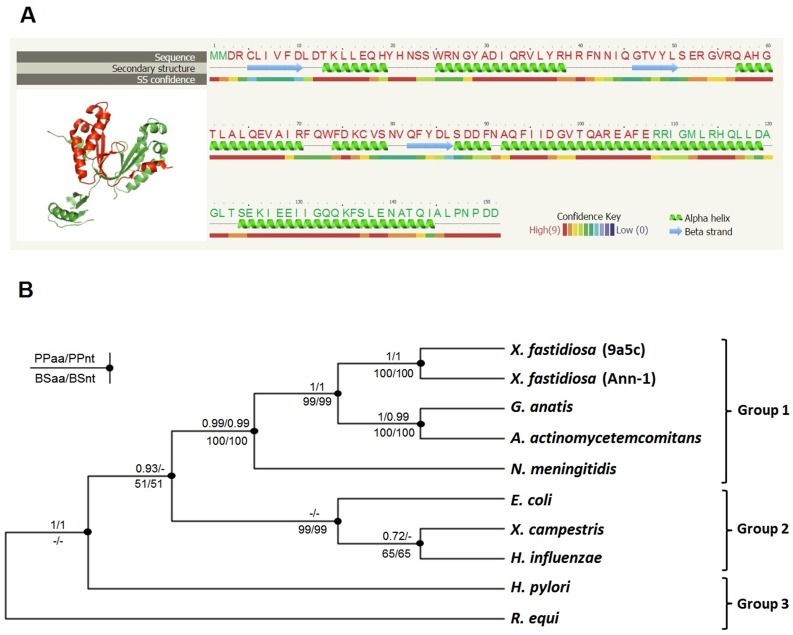
Fig 2. Structural and phylogenetic analysis of Xf-VapD.
(A) The prediction of the 3D structure of Xf-VapD was performed using Phyre V 2.0 (red cartoon). A model was obtained with 66% of the Xf-VapD sequence and 100% confidence using the single highest scoring template. The structure was edited using PyMol. On the right, the amino acid sequence is shown with the respective secondary structure and confidence. The amino acid residues highlighted in red correspond to the area in the red cartoon shown in the 3D structure. (B) The neighbor-joining consensus tree inferred for the amino acid sequences of VapD. The values above the branches indicate the Bayesian posterior probabilities for the amino acids (PPaa) and nucleotides (PPnt). The values below the branches indicate the results of the neighbor-joining method for the amino acids (BSaa) and nucleotides (BSnt) following 1,000 bootstrap replicates. The minus symbol (‒) indicates that no support was reached for this node. The groups are the same as those in Fig 1.