Fig 6. Twist1 expression elevated by binding of Sp1 transcription factor.
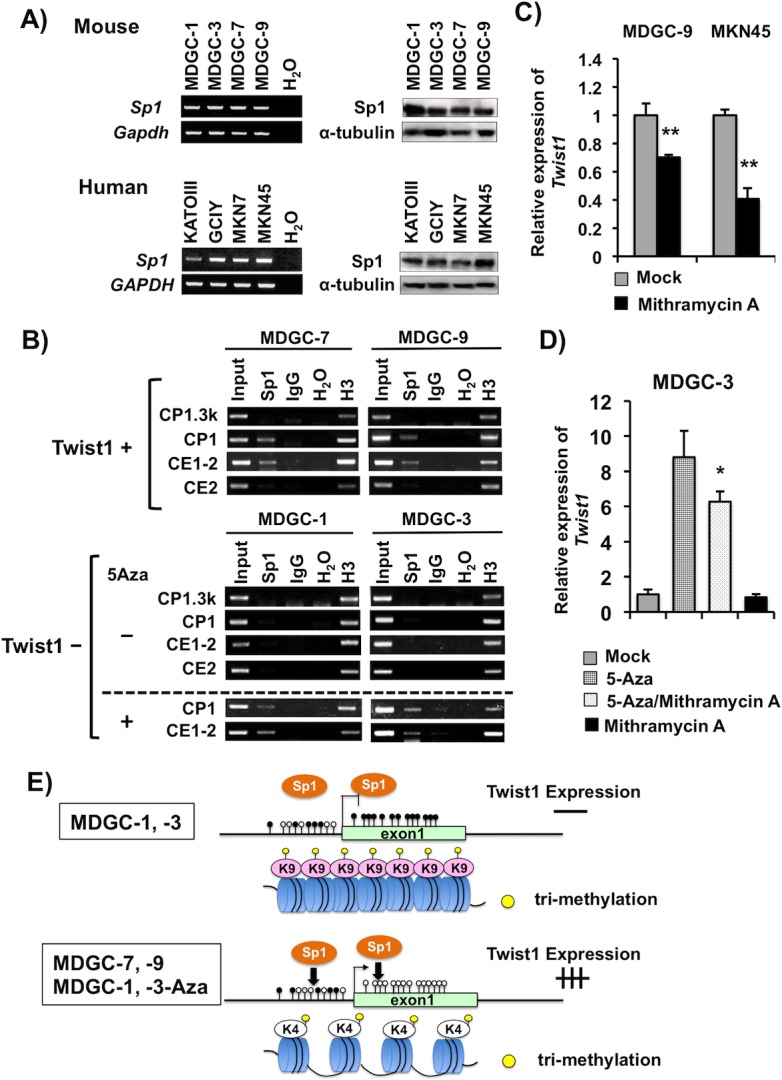
(A, B) RT-PCR (left) and Western blot (right) analyses of Sp1 mRNA and protein in MDGC cells and human GC cells. Gapdh and α-tubulin were used as internal controls for RT-PCR and Western blot, respectively. (B) ChIP assay showing the difference in Sp1 binding level between Twist1 expression-positive and -negative cells. Sp1 strongly bound to the CP1 and CE1-2 regions in Twist1 expression-positive cells (MDGC-7, MDGC-9). After 5-aza-dC treatment, Sp1 binding increased in Twist1 expression-negative cells (MDGC-1, MDGC-3). The regions analyzed are shown in Fig 2A. (C) Inhibition of Twist1 mRNA expression in murine and human GC cells after treatment with mithramycin A, a Sp1 binding inhibitor. (D) Alterations of Twist1 expression in MDGC-3 cells after treatment with 5-aza-dC, mithramycin A, and the combination. Twist1 expression was enhanced in MDGC-3 after treatment with 5-aza-dC but not mithramycin A. The Twist1 expression enhancement by 5-aza-dC was significantly inhibited by mithramycin A (*P<0.05). (E) Schematic representation of the transcriptional regulatory mechanisms of Twist1 in association with the epigenetic machinery and transcription factors. It is possible that Twist1 transcriptional activation is regulated through potential cooperation of DNA methylation, histone modification in complex with Sp1 binding to CpG-rich regions in exon 1.
