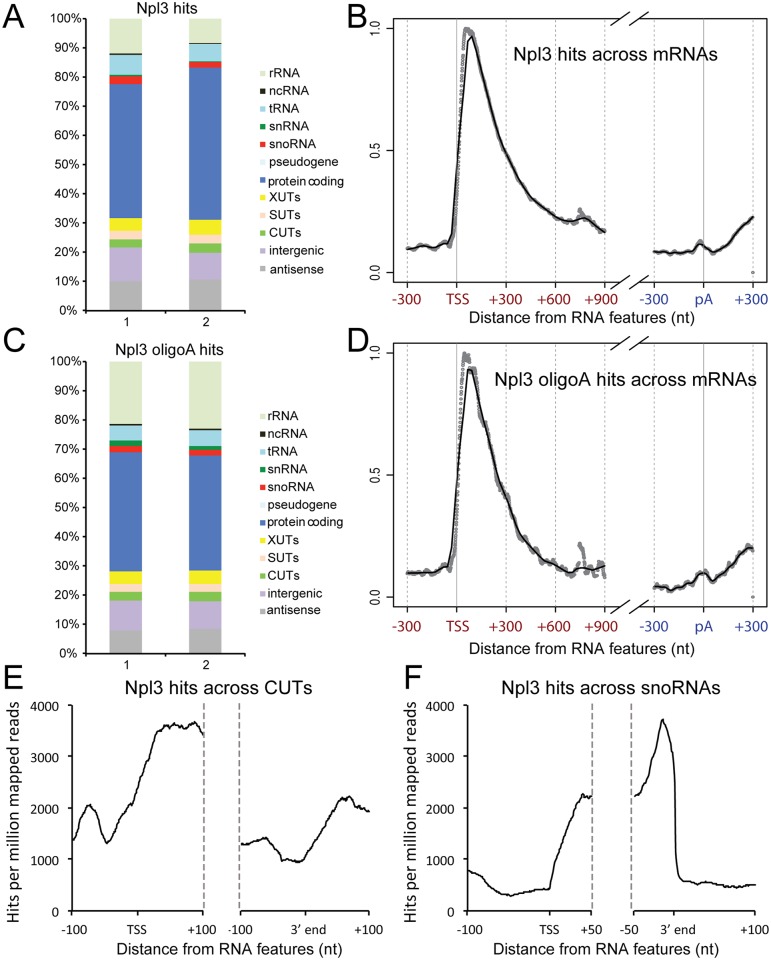
Fig 1. Distribution of Npl3 across RNAs is suggestive of a role in termination and surveillance.
A: Comparison of Npl3 binding across different RNA classes in two replicate CRAC experiments. Hits were assigned to RNA classes according to gene annotations from Ensembl (EF4.74) supplemented with additional features including ncRNAs, UTRs and antisense transcripts, as previously described [37]. Antisense, intergenic and ncRNA RNA classes are defined as follows. Antisense: a hit will be assigned as 'antisense' if it maps to regions of published antisense transcription [44,45] or to the opposite strand of a known feature. Intergenic: these are hits that map to any region of the genome where no other feature has been annotated. NcRNA: this category comprises 15 ncRNAs that don't fall into other categories, including scR1, SRG1 and TLC1. B: Distribution of Npl3 binding over mRNAs. Average distribution of Npl3 around the 5’ and 3’ ends of mRNAs. Transcripts are aligned at the transcription start sites (TSS) and polyA (pA) site. Grey dots depict precise number of hits at particular nucleotide positions and the dark green shows a line of best fit. Hits are normalized to a total of 1 across all mRNAs. C: Npl3 association with oligoadenylated mRNA fragments. As for panel A, but with data filtered for the presence of non-templated oligo(A) sequences (A2 or greater) recovered on target RNAs. D: Distribution of Npl3 binding over mRNAs. As for 1(B), but including reads containing oligoadenylated sequences only. E: Metagene analysis of Npl3 binding across CUTs. Distribution of Npl3 at all CUTs aligned by the TSS and 3' ends, with 100 nt flanks extending 100 nt into the 5' and 3' ends of transcripts. Only CUTs >150 nt in length were included in the analysis. F: Metagene analysis of Npl3 binding across snoRNAs. Distribution of Npl3 at all snoRNAs aligned by the TSS and 3' ends, with 100 nt flanks extending 50 nt into the 5' and 3' ends of transcripts.