Abstract
Francisella tularensis is a highly-infectious bacterium that causes the rapid, and often lethal disease, tularemia. Many studies have been performed to identify and characterize the virulence factors that F. tularensis uses to infect a wide variety of hosts and host cell types, evade immune defenses, and induce severe disease and death. This review focuses on the virulence factors that are present in the F. tularensis envelope, including capsule, LPS, outer membrane, periplasm, inner membrane, secretion systems, and various molecules in each of aforementioned sub-compartments. Whereas, no single bacterial molecule or molecular complex single-handedly controls F. tularensis virulence, we review here how diverse bacterial systems work in conjunction to subvert the immune system, attach to and invade host cells, alter phagosome/lysosome maturation pathways, replicate in host cells without being detected, inhibit apoptosis, and induce host cell death for bacterial release and infection of adjacent cells. Given that the F. tularensis envelope is the outermost layer of the bacterium, we highlight herein how many of these molecules directly interact with the host to promote infection and disease. These and future envelope studies are important to advance our collective understanding of F. tularensis virulence mechanisms and offer targets for future vaccine development efforts.
Keywords: Francisella, tularemia, tularensis, outer membrane proteins, virulence factors, envelope
Introduction
Francisella tularensis is a Gram-negative intracellular bacterium and the causative agent of the zoonotic disease tularemia (Carvalho et al., 2014). F. tularensis has been further subdivided into two subspecies: F. tularensis subsp. tularensis, also referred to as Type A strains which are found exclusively in North America; and F. tularensis subsp. holarctica, also referred to as Type B strains which are found throughout the northern hemisphere (Oyston et al., 2004; Keim et al., 2007). Type A and Type B strains are extremely infectious to humans and can be acquired through various routes, including arthropod bites, contact with infected animals, ingestion of contaminated food or water, or inhalation of aerosols (Kingry and Petersen, 2014). Depending on the route of infection, various organs and tissues are heavily colonized by F. tularensis, including skin, lymph nodes, lungs, spleen, liver, and kidney, with bacteremia common in the early stages of infection. Disease onset is typically very rapid with flu-like symptoms common, including fever, headache, chills, malaise, and sore throat (Dennis et al., 2001).
Because of its low infectious dose, multiple routes of infection, and high morbidity and mortality rates, F. tularensis has been designated as a Tier 1 Select Agent by the U.S. Centers for Disease Control and Prevention (CDC), highlighting concerns over its potential use a bioterrorism agent (Dennis et al., 2001). Type A strains are the most virulent (ID50 < 10 CFU via multiple infection routes in many animals, including humans; Ellis et al., 2002; Molins et al., 2010), with the human ulcer isolate, Schu S4, being the most commonly-used strain in BSL3 laboratories (Molins et al., 2014). Type B strains are highly-infectious to mice and guinea pigs (pulmonary and intradermal ID50 < 10 CFU; Ellis et al., 2002; Molins et al., 2010) but higher doses are needed to infect rabbits (106–109 CFU subcutaneously) and humans (< 103 via multiple routes; Ellis et al., 2002; Petersen and Molins, 2010). Despite these differences in virulence, Type B strains cause substantially more infections worldwide (Petersen and Molins, 2010; Hestvik et al., 2015). An attenuated Type B strain, the live vaccine strain (LVS), was developed in the former Soviet Union by serial passage through mice (Eigelsbach and Downs, 1961). Despite its name, LVS is not licensed for human vaccination in the U.S. due to safety and efficacy concerns (Oyston, 2009). However, LVS has been extensively used in research laboratories because of its ability to be safely used in BSL2 environments, high virulence in mouse models, high degree of genetic conservation with virulent Type A and Type B strains, and similar intracellular replication kinetics as virulent Type A and Type B strains (Elkins et al., 2007; Jones et al., 2014). A genetically-related species, Francisella novicida, also can be manipulated at BSL2 and does not cause disease in healthy humans, but F. novicida has a number of genetic and phenotypic differences that bring into question its use as a F. tularensis surrogate (Kingry and Petersen, 2014). The purpose of this review is to summarize the current knowledge on F. tularensis virulence factors. As such, we made every attempt throughout this review to clearly note what strain was used in each of the referenced studies so that readers can render their own judgments about the applicability to human disease.
Whereas, F. tularensis is a significant pathogen based on high morbidity and mortality rates, it also is an extremely interesting pathogen due its complicated intracellular lifestyle, ability to infect a wide variety of host cell types, persistence in the environment, and lack of classical bacterial virulence factors such as exotoxins or a Type III secretion system (Celli and Zahrt, 2013). Many excellent reviews previously have characterized F. tularensis as a “stealth” pathogen, which first evades host immune detection (Sjöstedt, 2006; Jones et al., 2014), but subsequently induces a cytokine storm that causes host death (Cowley, 2009; Cowley and Elkins, 2011). In addition, the metabolic pathways and nutrient requirements of F. tularensis promoting survival inside host cells also have been elegantly outlined (Barel and Charbit, 2013; Barel et al., 2015). Here, we will highlight studies that have identified and characterized more “classical” virulence factors of F. tularensis (i.e., those bacterial molecules that directly interact with the host, directly damage the host, or sense changes in the environment to modify bacterial gene expression). This review will begin with the outermost capsular layer and sequentially discuss the roles of LPS, the outer membrane, periplasm, and inner membrane in F. tularensis virulence and disease.
Capsule
Polysaccharide capsules are produced by bacteria such as Escherichia coli, Neisseria meningitidis, and Streptococcus pneumoniae (Preston and Dockrell, 2008; Willis and Whitfield, 2013), whereas protein capsules are produced by bacteria such as Bacillus anthracis (Cote et al., 2011). Capsule typically protects bacteria from complement-mediated lysis, phagocytosis, and immune recognition. For intracellular pathogens like F. tularensis, antiphagocytic properties ostensibly may appear counterproductive, but it is now well-appreciated that “stealth” pathogens manipulate host cell entry and intracellular trafficking mechanisms to limit inflammation and promote intracellular survival (Geier and Celli, 2011; Dai et al., 2013). Conversely, capsule can elicit protective immunity, as evidenced by successful capsule-based commercial vaccines against S. pneumoniae, Haemophilus influenza, and N. meningitidis (McIntyre et al., 2012). Given the disparate roles of bacterial capsule in virulence and protective immunity, Francisella capsule has been the focus of many research studies.
An electron-transparent, capsule-like outer structure, approximately 0.02–0.04 μm thick, was first described by Hood and shown to play a role in F. tularensis virulence (Hood, 1977). Capsule was observed in F. tularensis strain Schu S4 when bacteria were aerosolized or grown in modified casein hydrolysate liquid medium. However, capsule was absent from bacteria exposed to air long-term (20 h), treated with 10% sodium chloride, or taken from aged cultures (6 months at 4°C). Biochemical analyses indicated that F. tularensis capsule was distinct from cell wall material, being composed of ~51% lipids (primarily 14 and 16 carbon length fatty acids), up to 35% amino acids, and up to 21% carbohydrates (including mannose and rhamnose; Hood, 1977). Capsule production appears to be well-conserved among many F. tularensis strains, as a monoclonal antibody generated against Schu S4 capsular material cross-reacted with 14 different Type A and Type B isolates, including LVS, but did not react against LPS (Apicella et al., 2010). Detailed analysis of the F. tularensis capsule revealed polysaccharide identical to the O-antigen subunit of LPS that diffusely migrated on SDS-PAGE between 100 and 250 kDa (Apicella et al., 2010). As proof that F. tularensis capsule was composed of O-antigen and no other LPS components, mass spectrometry and NMR analyses demonstrated that purified capsule lacked other LPS-specific components, including 2-keto-3-deoxyoctulsonic acid (KDO) and lipid A (Apicella et al., 2010; Bandara et al., 2011). Given the large molecular weight and identical nature of F. tularensis capsule O-antigen and LPS O-antigen, F. tularensis capsule appears to be a Group 4 capsule (Whitfield, 2006). As noted above, whereas Schu S4 capsule was found to be composed of mannose and rhamnose, a LVS capsule-like complex was found to contain mannose, glucose, and galactose, indicating that capsular composition may differ between Type A and B species (Bandara et al., 2011). Together, these studies demonstrated that many F. tularensis strains express a polysaccharide capsule that it is identical to the O-antigen but lacks other LPS components.
Capsule has been shown to play an important role in F. tularensis virulence both in vitro and in vivo. For Schu S4, acapsular bacteria were attenuated when guinea pigs were infected intraperitoneally (i.p.) or when mice were infected either i.p. or intranasally (i.n.) (Hood, 1977; Lindemann et al., 2011; Rasmussen et al., 2014). In macrophages, Schu S4 acapsular mutants exhibited reduced cytosolic growth following phagosomal escape, due to the triggering of apoptotic or pyroptotic cell death (Lindemann et al., 2011). That study highlighted the importance of capsule to evade intracellular detection pathways. Capsule also contributes to LVS virulence, as serial passage on Chamberlain's medium increased LVS capsule production and capsulated LVS was substantially more virulent in mice via intravenous (i.v.), i.p., or i.n. routes (Cherwonogrodzky et al., 1994). Similarly, acapsular LVS mutants were found to be attenuated in macrophages and mice infected either i.p. or i.n. (Sandström et al., 1988; Bandara et al., 2011).
Whereas bacterial capsules typically block complement or antibody from binding to the bacterial surface, the role of F. tularensis capsule in serum resistance is less clear. In one study, chemically-mutagenized LVS rough colonies that lacked electron-dense surface layers (i.e., acapsular mutants) were susceptible to killing by non-immune human serum, primarily through the binding of IgM and complement component C3 (Sandström et al., 1988). A second study noted that F. tularensis spontaneous acapsular mutants absorbed more human serum components than wild-type capsulated strains and this effect primarily was mediated by IgM (Sorokin et al., 1996). Finally, Schu S4 acapsular mutants were found to be serum sensitive (Lindemann et al., 2011). While these three studies suggest that F. tularensis capsule blocks antibody and complement binding to the bacterial surface, other studies have suggested that serum resistance cannot be solely attributed to capsule. For example, an acapsular LVS mutant (ΔFTL1422–1423) was found to be completely resistant to both guinea pig and human serum (Bandara et al., 2011). In addition, a number of studies have noted antibody and/or complement binding to LPS (see Lipopolysaccharide Section). Based on these conflicting results, further studies are needed to better understand the role of capsule in serum resistance.
There has been much interest in identifying the genes involved in F. tularensis capsule production. However, given the above referenced studies noting that F. tularensis capsule and O-antigen polysaccharides are identical, it is not surprising that capsule gene identification studies have been difficult to interpret or have yielded conflicting results. A detailed analysis of gene deletion mutants indicated that wbtA1, wbtA2, wbtC, wbtI, wbtM, and FTL0708 were required for capsule production, whereas capB, capC, lpxL, wbtK, FTL0706, and FTT0673–FTT0674 were not (Apicella et al., 2010; see Table 1 for corresponding gene loci in Schu S4, LVS, or F. novicida). Of those genes, individual deletions of wbtA1, wbtA2, wbtC, wbtI, or wbtM resulted in loss of both capsule and O-antigen, highlighting that capsule and O-antigen share some biosynthetic pathway components. Conversely, only the FTL0708 mutant was found to be deficient in capsule production but not LPS, demonstrating that some genes may be capsule-specific. In a separate study, a LVS glycoprotein-containing capsule-like complex was linked to loci FTL1421–1432 and deletion of both FTL1422 and FTL1423 resulted in loss of this capsule-like complex (Bandara et al., 2011). In Schu S4, loci FTT1236–1238 was shown to be responsible for both O-antigen production and capsule biosynthesis, of which FTT1236 (waaY) and FTT1238 (waaL) were specifically required for Schu S4 capsule production (Lindemann et al., 2011). Whereas, the aforementioned studies focused on polysaccharide or O-antigen-related genes, a three gene locus named capBCA, homologous to the B. anthracis poly-γ-D-glutamic acid capsule cap locus, was found to be required for LVS i.n. virulence in mice (Su et al., 2007). The capBCA locus is conserved across Type A and B strains, and mutagenesis studies in Schu S4 and LVS confirmed that capBCA or capB mutants were attenuated both in vitro and in vivo (avirulent by subcutaneous [sub-q], intradermal [i.d.], and i.n. routes; Su et al., 2007; Jia et al., 2010; Michell et al., 2010). However, capsule production was not assessed in any of those virulence studies, bringing into question if the capBCA locus truly is involved in F. tularensis capsule production. Indeed, as noted earlier, Apicella et al., found that capB and capC were not involved in LVS capsule production, suggesting that the cap genes perform another function. A more recent study suggested that capA may encode an integral inner membrane protein that interacts with CapB and CapC to export carbohydrates extracellularly (Martin-Garcia et al., 2014). Given the large number of genes and operons that have been reported to play roles in F. tularensis capsule production, more studies are clearly needed.
Table 1.
Francisella genes involved in capsule and/or LPS synthesis.
| Gene name (if designated) | Schu S4 locus | LVS locus | F. novicida strain U112 locus | |
|---|---|---|---|---|
| lpxD2 | FTT0286c | FTL0196 | FTN0200 | LPS |
| flmF2 | FTT0454 | FTL1611 | FTN0545 | LPS |
| flmK | FTT0455c | FTL1609 | FTN0546 | LPS |
| FTT0789 | FTL1432 | FTN1221 | Capsule | |
| FTT0790 | FTL1431 | FTN1220 | Capsule | |
| FTT0791 | FTL1430 | FTN1219 | Capsule | |
| FTT0792 | FTL1429 | FTN1218 | Capsule | |
| FTT0793 | FTL1428 | FTN1217 | Capsule | |
| FTT0794 | FTL1427 | FTN1216 | Capsule | |
| FTT0795 | FTL1426 | FTN1215 | Capsule | |
| FTT0796 | FTL1425 | FTN1214 | Capsule | |
| FTT0797 | FTL1424 | FTN1213 | Capsule | |
| FTT0798 | FTL1423 | FTN1212 | Capsule | |
| FTT0799 | FTL1422 | FTN1211 | Capsule | |
| FTT0800 | FTL1421 | FTN1210 | Capsule | |
| capA | FTT0805 | FTL1416 | FTN1201 | unknown |
| capC | FTT0806 | FTL1415 | FTN1200 | unknown |
| capB | FTT0807 | FTL1414 | FTN1199 | unknown |
| lpxE | FTT0891 | FTL0393 | FTN0416 | LPS |
| waaY | FTT1236 | FTL0708 | FTN1254 | Capsule |
| waaZ | FTT1237 | FTL0707 | FTN1255 | LPS |
| waaL | FTT1238 | FTL0706 | FTN1256 | Capsule |
| wbtM | FTT1450c | FTL0606 | FTN1416 | Both |
| wzx | FTT1453c | FTL0603 | FTN1419 | LPS |
| wbtI | FTT1455c | FTL0601 | FTN1421 | Both |
| wzy | FTT1458c | FTL0598 | FTN1424 | LPS |
| wbtF | FTT1459c | FTL0597 | FTN1425 | LPS |
| wbtE | FTT1460c | FTL0596 | FTN1426 | LPS |
| wbtD | FTT1461c | FTL0595 | FTN1427 | LPS |
| wbtC | FTT1462c | FTL0594 | FTN1428 | Both |
| wbtA2 | FTT1463c | FTL0593 | FTN1430 | Both |
| wbtA1 | FTT1464c | FTL0592 | FTN1431 | Both |
| kdtA | FTT1561 | FTL0547 | FTN1469 | LPS |
| lpxD1 | FTT1571c | FTL0537 | FTN1480 | LPS |
| lpxF | FTT1643c | FTL1815 | FTN0286 | LPS |
The surface localization of bacterial capsules often has made them attractive targets for vaccine development efforts. However, Francisella capsular material has yielded mixed results as a protective antigen. Schu S4 capsular material did not elicit protection in guinea pigs or mice against i.p. Schu S4 challenge (Hood, 1977). However, immunization with LVS capsular material protected mice from i.p. LVS challenge, despite not protecting mice from i.p. Schu S4 challenge (Apicella et al., 2010). The use of capsule-deficient strains as live attenuated vaccines has yielded more promising results, with a LVS ΔFTL1422–1423 strain protecting against high-dose wild-type i.n. LVS challenge (Bandara et al., 2011) and Schu S4 ΔwaaY and ΔwaaL strains protecting against wild-type Schu S4 i.p. and i.n. challenge (Rasmussen et al., 2014). Importantly, the use of capsule-deficient mutants as vaccine strains indicates that F. tularensis capsule may not be a protective antigen but that capsule-deficient mutants are highly-attenuated and could be safe and effective vaccine candidates (Bandara et al., 2011). Despite the unlikely role of the capBCA locus in capsule production, a LVS capBCA mutant protected mice from wild-type i.n. LVS challenge (Su et al., 2007). Similarly, both LVS and Schu S4 ΔcapB mutants protected mice against Schu S4 aerosol and sub-q challenge (Jia et al., 2010; Michell et al., 2010).
In summary, a majority of studies have demonstrated that F. tularensis produces a polysaccharide capsule that is identical to the O-antigen and that capsule is required for F. tularensis virulence (Figure 1). Despite a number of studies demonstrating that capsule is required for serum resistance, some controversy remains about the true role of capsule in serum resistance, specific composition of the F. tularensis capsule, and the genes required for capsule production. In addition, some aberrant findings suggest that multiple capsule-like substances may be produced by different Francisella strains and that capsule composition might vary depending on culture conditions or environmental stimuli. If various capsule compositions do indeed exist, these capsule variations may be an important part of the overall virulence strategy of Francisella.
Figure 1.
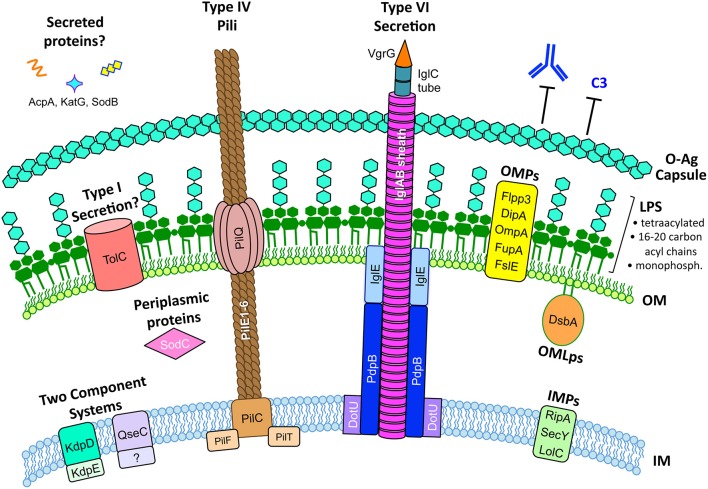
The F. tularensis envelope and its role in virulence. The F. tularensis capsule, identical to the O-antigen of LPS, blocks IgM and complement component C3 binding. F. tularensis LPS differs from prototypic LPS in that it is tetraacylated, possesses long-chain acyl chains (16–20 carbons long), is either non- or mono-phosphorylated, and is a weak TLR4 agonist. Type IV pili form horn-like protrusions on the bacterial surface and, while correlated with virulence, the exact function of F. tularensis type IV pili are not known. A Francisella Pathogenicity Island (FPI)-encoded Type VI-like secretion system has been reported in F. novicida (Table 3) but expression and function in virulent F. tularensis strains has not been reported. Various proteins have been reported to be secreted by F. tularensis, including AcpA, KatG, and SodB (Table 3), but the mechanisms for their secretion are unknown. The F. tularensis outer membrane (OM) contains many outer membrane proteins (OMPs) and outer membrane lipoproteins (OMLps) that are required for virulence (Table 4). Periplasmic and inner membrane proteins (IMPs) also play roles in F. tularensis virulence (Table 5), but much less is known about these envelope compartments. See individual sections for specific details and references.
Lipopolysaccharide
Lipopolysaccharide (LPS) makes up the outer leaflet of the outer membrane for most Gram-negative bacteria. LPS is composed of three main components; lipid A, core oligosaccharide, and the O-antigen (Miller et al., 2005). Of these, lipid A (endotoxin) is highly immunostimulatory and lipid A modifications are a common virulence strategy for many Gram-negative pathogens (Tan and Kagan, 2014). E. coli lipid A is the prototype for many Gram-negative lipid A structures, with six acyl chains (hexaacylated) that are 12–14 carbons in length. By comparison, the atypical lipid A of Francisella is tetraacylated with fatty acid chains 16–18 carbons in length (Vinogradov et al., 2002; Phillips et al., 2004). These F. tularensis LPS modifications, and others described below, greatly reduce TLR4 activation and allow for immune evasion.
A long-recognized F. tularensis blue-gray colony variation has been shown to correlate with differences in virulence, host cell stimulation, immunogenicity, and alterations in LPS (Eigelsbach et al., 1951; Hartley et al., 2006; Soni et al., 2010). In fact, the highly-virulent Type A strain Schu S4 owes its “S4” designation to the “S4” smooth blue colony phenotype, as compared to the “S1” and “S2” smooth buff-colored colony phenotypes, or “NS” non-smooth gray colony phenotypes that were isolated from this clinical isolate (Eigelsbach et al., 1951). In i.p. mouse infection studies, smooth blue Schu S4 variants were extremely virulent whereas non-smooth gray Schu variants were six-logs attenuated (Eigelsbach et al., 1951). Gray variants also have been reported in LVS and these spontaneously-occurring colony variants were attenuated in rat macrophages due to their induction of high levels of nitric oxide (NO), as compared with wild-type LVS which replicated to high numbers in rat macrophages and induced very little NO (Cowley et al., 1996). Differences in virulence and macrophage activation appeared to be due to alterations in lipid A, as purified lipid A from the LVS gray variant induced more NO than lipid A from wild-type LVS. In that same study, F. novicida whole bacteria or purified lipid A from F. novicida were found to induce more NO than either wild-type LVS bacteria or gray variant LVS bacteria, demonstrating that F. novicida LPS is distinct from F. tularensis LPS (Cowley et al., 1996). More recent work on gray variant LVS determined two key features: first, gray variants express less LPS O-antigen than wild-type LVS; second, gray variants have reduced lipid A galactosamine modifications compared to wild-type LVS. Glycosyltransferase genes flmF2 (FTL_1611) and flmK (FTL_1609) were found to be expressed at significantly lower levels in gray-variant LVS and deletion of flmF2 from wild-type LVS reduced the frequency of phase variation, supporting a role for these glycosyltransferases in phase variation (Soni et al., 2010). Finally, whereas wild-type LVS protected mice against Schu S4 i.n. challenge, LVS gray variants could not, indicating that O-antigen expression levels and lipid A galactosamine modifications likely are important for immunogenicity.
Detailed NMR and mass spectrometry studies have revealed that LPS O-antigen polysaccharide repeats are identical between Schu S4 and LVS, but differ from F. novicida LPS (Prior et al., 2003; Vinogradov and Perry, 2004; Thomas et al., 2007; Wang et al., 2011). The differences in LPS O-antigen are believed to be due to genomic differences in the wzy, wzx, and wbt loci, with Schu S4 and LVS both containing 15 O-antigen genes, as compared to F. novicida which contains 12 O-antigen genes (Prior et al., 2003; Thomas et al., 2007). For all Francisella strains, the loci are flanked by transposases and have a lower G-C content than the rest of the Francisella genome, suggesting possible horizontal gene acquisition of the O-antigen operon. Deletion of three highly-conserved LPS O-antigen genes, wbtDEF, in F. novicida and F. tularensis Schu S4 resulted in attenuation in mice via a sub-q infection route and loss of LPS O-antigen production in both bacterial species. However, despite the mutual loss of LPS O-antigen, only F. novicida wbtDEF mutants were serum sensitive and only Schu S4 wbtDEF mutants were attenuated for growth in mouse macrophages (Thomas et al., 2007). In contrast, whereas LVS wbtDEF mutants also were found to be LPS O-antigen deficient and serum sensitive, LVS wbtDEF mutants were not attenuated for human macrophage replication (Clemens et al., 2012). Taken together, these results indicate that although the Francisella LPS O-antigen is generally required for virulence in animals, the LPS O-antigen may play subtly different roles in intracellular survival and serum resistance among the different Francisella species. Importantly, LPS O-antigen composition appears to be critical for protective immunity, as F. novicida LPS protects against i.p. F. novicida challenge, LVS LPS protects against sub-q LVS challenge, and Schu S4 LPS protects against sub-q LVS challenge. In contast, F. novicida LPS does not protect against sub-q LVS or sub-q Schu S4 challenge and Schu S4 LPS does not protect against i.p. F. novicida challenge (Fulop et al., 2001; Thomas et al., 2007). The importance of LPS O-antigen in stimulating protective immune responses was further highlighted by the fact that vaccination with a F. novicida wbtDEF mutant was unable to protect mice from wild-type F. novicida i.p. challenge and a SchuS4 wbtDEF mutant was unable to protect mice from wild-type Schu S4 sub-q challenge (Thomas et al., 2007). Follow-up studies have confirmed that Schu S4 wbtC and wbtI also play roles in LPS O-antigen synthesis, as both mutants had altered or missing LPS O-antigen, were attenuated in mice infected i.d., and did not protect against low dose Schu S4 aerosol challenge. In addition to its role in LPS O-antigen synthesis, wbtC also appears to play a role in protein glycosylation, as wbtC mutants displayed fewer glycosylated isoforms of both DsbA (see Outer Membrane Proteins Section) and PilA (see Type IV Pili Section), which previously were reported to be glycosylated in F. tularensis (Twine et al., 2012).
In most Gram-negative bacteria, lipid A is linked to the core region through two KDO molecules (Okan and Kasper, 2013). By comparison, the F. tularensis core region contains only one KDO unit (Vinogradov et al., 2002; Vinogradov and Perry, 2004). Deletion of the putative core oligosaccharide KDO transferase gene, kdtA, from Schu S4 resulted in loss of the core oligosaccharide. Although, kdtA mutants still produced O-antigen, it was not present on the bacterial surface, supporting the role of kdtA in core oligosaccharide synthesis. In agreement with other studies demonstrating the importance of LPS in virulence and immunogenicity, Schu S4 kdtA mutants were avirulent in mice via an i.d. infection route but were not protective against Schu S4 aerosol challenge (Twine et al., 2012).
The lipid A component of Francisella LPS is conserved across species but possesses key differences from conventional LPS. First, Francisella lipid A is tetraacylated, as compared to the hexaacylated lipid A commonly found in most Gram-negatives. Second, Francisella lipid A acyl chains are 16 to 18 carbons long, as compared to the 12–14 carbon length chains of conventional LPS. Third, Francisella lipid A is either non-phosphorylated or mono-phosphorylated (at the 1′ position), as compared to prototypic lipid A which is phosphorylated at the 1′ and 4′ positions of the diglucosamine backbone. Fourth, Francisella masks its 1′ phosphate, if present, by adding galactosamine. The reduction or absence of negatively-charged phosphates likely affects the overall charge of Francisella LPS and confers resistance to antimicrobial peptides (Gunn and Ernst, 2007; Okan and Kasper, 2013). More comprehensive analyses of Francisella lipid A have revealed that a variety of structures exist, including acyl chains of varying lengths (from 14 to 20 carbons) and various sugar substitutions at the 1′ and 4′ backbone positions (Schilling et al., 2007; Beasley et al., 2012). Orthologs of the nine E. coli lipid A synthesis enzymes exist in F. tularensis, indicating that nascent Francisella lipid A is synthesized with phosphate groups at both 1′ and 4′ positions. However, Francisella actively dephosphorylates its lipid A at the 1′ position using the LpxE phosphatase and at the 4′ position using the LpxF phosphatase (Wang et al., 2004, 2006). The importance of lipid A desphosphorylation as a virulence mechanism is highlighted by studies demonstrating that F. novicida lpxF mutants are avirulent in macrophages and mice (aerosol, i.d., and sub-q infection routes tested) and are hypersensitive to antimicrobial peptides (Wang et al., 2007; Kanistanon et al., 2012). As noted above, LPS appears to generate species-specific immune responses, as the F. novicida ΔlpxF strain protected mice against wild-type i.p. or aerosol F. novicida challenge but did not confer protection against virulent Type A or Type B challenge (Kanistanon et al., 2012). Francisella lipid A also has been reported to be unique from other Gram-negative lipid A molecules in that 60–95% can be expressed as “free” lipid A (without core-oligosaccharide or O-antigen) in LVS and F. novicida (Wang et al., 2006; Lai et al., 2010; Barker et al., 2014). Whereas, Yersinia pestis has been shown to alter the number of acyl chains at different temperatures (Rebeil et al., 2006), Francisella modifies its acyl chain length based on temperature changes. At environmental temperatures (18°C), the F. novicida LpxD2 enzyme adds 16 carbon-length acyl chains to lipid A, whereas at mammalian temperatures (37°C), the F. novicida LpxD1 enzyme adds 18 carbon-length acyl chains. These acyl chain length modifications appear to be important for virulence as a F. novicida lpxD1 mutant was attenuated in mice via the sub-q route (Li et al., 2012). It has been speculated that these lipid A modifications alter the permeability and fluidity of the outer membrane and may affect outer membrane protein distributions or interactions, which may help Francisella adapt to survival in insects and mammalian hosts. Although lpxD1 and lpxD2 genes exist in Type A and Type B strains, it is unclear if Type A and Type B strains perform similar carbon chain length modifications based on temperature.
All of the above modifications appear to be important for Francisella LPS to minimize recognition by LPS recognition molecules, including LPS-binding protein, MD-2, and TLR4 (Ancuta et al., 1996; Barker et al., 2006; Hajjar et al., 2006). Conventional LPS (i.e., E. coli) typically binds to LPS recognition proteins and induces B-cell proliferation. However, only very high doses (~75 μg) of F. novicida LPS were able to induce mouse splenocyte proliferation, demonstrating its weak B-cell mitogen activity. Interestingly, similar amounts of LVS LPS did not induce any splenocyte proliferation (Kieffer et al., 2003). As compared to E. coli LPS, LVS LPS was not found to be endotoxic by either the Limulus amoebocyte assay or in mice sensitivity studies (Sandström et al., 1992). Whereas, monocyes and macrophages produced little to no cytokines in response to LVS LPS, F. novicida LPS stimulated macrophages to produce IL-12 and TNFα, highlighting that F. novicida is more pro-inflammatory than F. tularensis (Sandström et al., 1992; Kieffer et al., 2003).
Despite LPS modifications to avoid various innate immune pathways, LVS LPS was reported to activate the classical complement pathway (Fulop et al., 1993). Complement activation is a double-edged sword for F. tularensis, as C3b opsonization is important for host cell phagocytosis but the bacteria must avoid formation of the membrane attack complex. Follow-up studies confirmed that complement component C3 deposits on Schu S4, LVS, and F. novicida, but all three strains were complement resistant due to rapid inactivation of complement (conversion to C3bi), which was dependent on the presence of O-antigen (Clay et al., 2008). F. tularensis LPS-complement interactions also appear to play a role in macrophage uptake as, in the presence of complement, wild-type LVS was phagocytosed by spacious pseudopod loops, whereas LVS ΔwbtDEF or wbtIG191V O-antigen mutants were phagocytosed by tight overlapping pseudopod layers (Clemens et al., 2012). As noted above, since the capsule and O-antigen may share some biosynthetic pathway components, these results are difficult to interpret. However, these results do suggest that O-antigen components, either in the capsule or LPS, may block complement-complement receptor interactions, leading to altered macrophage uptake mechanisms.
Taken together, Francisella species have evolved distinct LPS molecules, with a number of structural variations from prototypic LPS, which promote immune evasion. The inert nature of Francisella LPS is particularly true for Type A and Type B strains. Further, many of the outlined studies demonstrate that specific LPS modifications contribute to the overall virulence strategy of F. tularensis, resulting in high infectivity and severe disease. Despite the low stimulatory capacity of F. tularensis LPS, it is interesting to note that this molecule provides species-specific protection. As such, future vaccine development efforts may need to consider incorporating anti-LPS responses that cross-react with both Type A and Type B strains.
Type IV pili
Type IV pili are important virulence factors for many Gram-negative pathogens, having been shown to play important roles in bacterial adhesion, aggregation, twitching motility, and DNA uptake (Craig et al., 2004; Berry and Pelicic, 2015). F. tularensis type IV pili first were reported in 2004 as “horn-like” surface structures that were observed when LVS was grown on agar medium. In contrast, long thin fibers were observed when LVS was grown in liquid medium, indicating that pili-like structures may phenotypically vary depending on culture conditions (Gil et al., 2004). Type IV pili expression also has been reported in Schu S4 and F. novicida (Zogaj et al., 2008; Ark and Mann, 2011), suggesting that type IV pili are expressed in other Francisella species. However, there also are indications that pilus expression may be strain- or lab-dependent, as two publications have noted that pilus-like structures were not observed on a virulent F. tularensis subsp. holarctica strain or in F. novicida (Forslund et al., 2006; Hager et al., 2006).
Genomic sequencing has confirmed the existence of gene clusters in various Francisella species with homology to the type IV pilus genes of Neisseria and Pseudomonas (Gil et al., 2004; Larsson et al., 2005). The Francisella type IV pilus gene clusters appear to contain 14 genes, including various outer membrane, inner membrane, ATPase, and pilin subunit proteins. The surface fiber is composed of between five and six pilin subunits, PilE1–PilE6 (Gil et al., 2004; Ark and Mann, 2011). Whereas all six putative pilin genes are present in Type A strains and F. novicida, Type B strains contain various truncations or deletions of pilE1, pilE2, or pilE3 (Ark and Mann, 2011). Indeed, LVS is missing pilE1 and premature stop codons have been introduced into pilE2 and pilE3 (Gil et al., 2004). By comparison, virulent Type B PilE1 (previously described as PilA) was found to form multimers at the bacterial surface and was extensively glycosylated during animal infections, suggesting that type IV pilin glycosylation plays an important role in virulence (Forslund et al., 2006). PilO (also referred to as PglA) was shown to be responsible for PilE1 glycosylation (Egge-Jacobsen et al., 2011). PilE1 has been noted to be required for full virulence of SchuS4 and a virulent Type B strain in sub-q mice infections (Forslund et al., 2006, 2010). The role of PilE1 in F. tularensis virulence is further supported by work demonstrating that restoration of pilE1, either alone or in combination with FTT0918 (described in Outer Membrane Proteins Section), increases LVS virulence (Salomonsson et al., 2009). Sequence analysis revealed that low-virulence Francisella strains contain a long pilE4 gene (F. novicida encodes 316 amino acids; F. tularensis LVS encodes 301 amino acids), whereas high-virulence strains contain a truncated pilE4 gene (subsp. tularensis encodes 197 amino acids; subsp. holarctica encodes 211 amino acids; Zogaj et al., 2008). Despite these differences in amino acid length, PilE4 appears to be the major pilin component and was required for type IV pilus assembly in Schu S4, LVS, and F. novicida (Zogaj et al., 2008; Ark and Mann, 2011). Interestingly, although PilE4 was required for LVS virulence, it was not required for Schu S4 virulence in mice via either sub-q or i.d. infection routes (Ark and Mann, 2011). However, as noted above, the reason for this virulence difference may be due to truncated pilE4 in Schu S4. The roles of PilE5 and PilE6 in type IV pili assembly and virulence are less clear, as neither were required for pili-like fiber assembly or Schu S4 virulence by either sub-q or i.d. infection routes but both contributed to LVS virulence by both sub-q and i.d. infection routes (Ark and Mann, 2011).
In addition to the surface-exposed pilin subunits, many type IV pili assembly proteins also are required for Francisella virulence. Deletion of either pilC, an inner membrane protein, or pilQ, an outer membrane protein (Huntley et al., 2007), resulted in Schu S4 attenuation in sub-q mice infections (Forslund et al., 2010). Additionally, PilF, the pilus assembly ATPase, and PilT, the pilus retraction ATPase, both were found to be required for LVS virulence in i.d. mice infections (Chakraborty et al., 2008). Interestingly, PilT was not required for Schu S4 virulence in sub-q mice infection studies (Forslund et al., 2010).
In F. novicida, individual components of the type IV pili homologous genes have been reported to be involved in a type II-like protein secretion system (Hager et al., 2006; Zogaj et al., 2008). Seven proteins were reported to be secreted by this putative F. novicida secretion system, including two chitinases (ChiA and ChiB), a chitin binding protein (CbpA), a protease (PepO), a beta-glucosidase (BglX), and two hypothetical proteins (Fsp58 and Fsp53; Hager et al., 2006). Although a type II-like secretion system have not been reported in virulent F. tularensis strains, it is interesting to speculate that the chitinases and CbpA may be important for survival or replication in ticks or other arthropod vectors. In addition, PepO and BglX were found to be regulated by the master virulence regulator MglA, suggesting that they may play roles in damaging host cells (Hager et al., 2006). However, the absence of pepO in virulent Type A and Type B F. tularensis strains suggests that these findings may be unique to F. novicida.
In summary, there are a number of species-specific variations in type IV pili genes, media-dependent variations in type IV pili expression, and conflicting reports about the roles of type IV pili in F. tularensis virulence. As such, further studies are needed to better understand F. tularensis type IV pili (summarized in Table 2 and Figure 1).
Table 2.
Francisella genes associated with type IV pili.
| Gene name | Schu S4 locus | LVS locus | F. novicida strain U112 locus | Function/Location |
|---|---|---|---|---|
| pilC | FTT1134 | FTL0827 | FTN1116 | Inner membrane |
| pilE1 (pilA) | FTT0890c | absent | FTN0415 | Pilin subunit |
| pilE2 (pilE) | FTT0889c | FTL0391 | FTN0414 | Pilin subunit |
| pilE3 (pilV) | FTT0888c | FTL0390 | FTN0413 | Pilin subunit |
| pilE4 | FTT0861c | FTL0359 | FTN0389 | Pilin subunit |
| pilE5 | FTT0230c | FTL0181 | FTN0070 | Pilin subunit |
| pilE6 | FTT1314c | FTL1475 | FTN0664 | Pilin subunit |
| pilF | FTT1133 | FTL0828 | FTN1115 | Assembly ATPase |
| pilO (pglA) | FTT0905 | FTL0425 | FTN1139 | PilE1 glycosylase |
| pilQ | FTT1156c | FTL0800 | FTN1137 | Outer membrane |
| pilT | FTT0088 | FTL1771a, FTL1770a |
FTN1622 | Retraction ATPase |
| chiA | FTT0715 | FTL1521 | FTN0627 | Secreted proteinb |
| chiB | FTT1786c | FTL0093 | FTN1744 | Secreted proteinb |
| cbpA | FTT1577 | FTL0532 | FTN1485 | Secreted proteinb |
| pepO | absent | absent | FTN1186 | Secreted proteinb |
| bglX | FTT1069c | FTL0543 | FTN1474 | Secreted proteinb |
| fsp58 | FTT0580 | FTL1331 | FTN0753 | Secreted proteinb |
| fsp53 | FTT1330 | FTL1491 | FTN0647 | Secreted proteinb |
A nonsense mutation in the LVS PilT coding sequence has resulted two loci, FTL1771 and FTL1770.
In F. novicida, individual components of the type IV pili system were reported to be involved in a type II-like secretion system. See text for more details.
Secretion systems and secreted proteins
Type I secretion
Type I secretion systems contribute to bacterial virulence through multi-drug efflux and toxin secretion, typified by the hemolysin secretion system of E. coli or TolC secretion system in many Gram-negative bacteria (Piddock, 2006; Thomas et al., 2014; Costa et al., 2015). F. tularensis contains two TolC orthologs, TolC and FtlC, both of which are outer membrane-localized (Huntley et al., 2007). Deletion of either tolC or ftlC from LVS increased sensitivity to antibiotics, dyes, and detergents, suggesting that TolC and FtlC play important roles in F. tularensis drug resistance (Gil et al., 2006). Additionally, TolC, but not FtlC, was required for LVS i.d. virulence in mice, indicating that these two TolC orthologs perform non-redundant functions in F. tularensis (Gil et al., 2006). A later study found that TolC marginally contributes to Schu S4 i.d. virulence, as tolC mutants resulted in 2 day-delayed time-to-death in mice, compared with wild-type infected mice (Kadzhaev et al., 2009). It remains possible that TolC may secrete yet-to-be discovered virulence factors, as LVS tolC mutants exhibited in vivo dissemination and survival defects, were unable to suppress pro-inflammatory cytokines, and could not inhibit macrophage apoptosis (Platz et al., 2010; Doyle et al., 2014). Another F. tularensis outer membrane protein, SilC, originally named due to homology with silver cation efflux proteins in other bacteria, shares sequence homology with F. tularensis TolC and FtlC and other bacterial TolC orthologs (Huntley et al., 2007). However, the role of SilC in F. tularensis secretion or virulence is unknown.
Type III and type IV secretion
Type III and Type IV bacterial protein secretion systems are well-known to inject bacterial effectors into host cells, either across the plasma membrane or across internal membranes, promoting bacterial infection and disease (Raymond et al., 2013; Burkinshaw and Strynadka, 2014; Christie et al., 2014). Thus far, functional Type III or Type IV protein secretion systems have not been described in F. tularensis and Type III or Type IV secretion system orthologs were not identified in the Schu S4 genome (Larsson et al., 2005). However, subsequent comparative genomic analysis found putative Type III effector and Type IV secretion system homologs in all species of Francisella. Interestingly, three of the Type III effector homologs and two of the Type IV secretion system homologs only were present in the avirulent strains (e.g., F. novicida and F. philomiragia) and were either truncated or absent from Type A, Type B, and F. tularensis subsp. mediasiatica strains, suggesting that these Type III and Type IV secretion system components may not contribute to human virulence (Champion et al., 2009).
Type VI secretion
Type VI secretion systems are related to T4-like bacteriophage injection systems and allow translocation of various effector proteins into host or competing bacterial cells (Ho et al., 2014; Russell et al., 2014). Whereas, a putative Type VI protein secretion system reportedly is encoded by the Francisella Pathogenicity Island (FPI), F. novicida has been almost exclusively used in these descriptions (de Bruin et al., 2007; Barker et al., 2009). The F. novicida Type VI secretion system appears to be composed of an outer sheath made up of IglA/IglB heterodimers arranged in a helical cylinder. IglA/IglB sheath assembly was shown to be initiated by various environmental signals, including macrophage uptake, 5% KCl, and altered oxygen tension (Clemens et al., 2015). Structural modeling has suggested that IglC forms the inner tube of the Type VI secretion system and contraction of the sheath has been theorized to drive the inner tube into target membranes (de Bruin et al., 2011; Clemens et al., 2015). VgrG has been suggested to localize to the tip of the Type VI secretion system, forming a trimeric host cell puncturing device (Bröms et al., 2012a). The sheath and tube complex presumably is anchored to the outer membrane by the outer membrane lipoprotein IglE (Robertson et al., 2013; Nguyen et al., 2014). Interestingly, IglE has been shown to interact with the PdpB and DotU inner membrane protein complex and studies suggest that IglE and PdpB form a periplasm-spanning channel (de Bruin et al., 2011; Nguyen et al., 2014). As expected, each of these Type VI structural components have been reported to play important roles in virulence, with IglA and IglB being required for F. novicida phagosomal escape, intracellular replication, and virulence in chicken embryo and mouse i.d. infection models (de Bruin et al., 2007; Bröms et al., 2009; Clemens et al., 2015), DotU and VgrG were necessary for LVS or F. novicida phagosomal escape and lethal i.d. or i.n. infections in mice (Barker et al., 2009; Bröms et al., 2012a), and IglE was required for F. novicida replication in macrophages and lethal i.n. infections in mice (Nguyen et al., 2014).
Despite the above studies noting the existence of a F. novicida Type VI secretion system and its role in virulence, the exact secreted effectors are still controversial. In one study, IglC,E,F,I,J, PdpA,E, and VgrG were found to be secreted by LVS into macrophages in a Type VI secretion-dependent manner. However, in that same study, F. novicida only was found to secrete IglC,E and PdpA,E indicating that there may be species-specific differences in Type VI secreted effectors (Bröms et al., 2012b). In contrast, a separate study reported that F. novicida secreted IglA-J, PdpA,C,E, DotU, and VgrG during macrophage infections (Hare and Hueffer, 2014). Indeed, the role of VgrG in Francisella is extremely unclear, with studies indicating that VgrG is a secreted effector (Barker et al., 2009; Clemens et al., 2015), coordinates secretion of other effector molecules (Barker et al., 2009), and/or is a structural component of the Type VI secretion apparatus (de Bruin et al., 2011; Bröms et al., 2012a). Understudied FPI and/or Type VI secretion system components include: IglG and IglI, which were required for LVS phagosomal escape and in vivo dissemination (Bröms et al., 2011); and pdpD and anmK, which were required for F. novicida virulence in chicken embryos and mice (Ludu et al., 2008). Interestingly, although anmK (predicted chaperone) is found in F. novicida, anmK is separated into two smaller ORFs in Type A F. tularensis strains, and anmK is absent from Type B strains, suggesting that anmK is not required for virulence in pathogenic F. tularensis strains (Bröms et al., 2010). Additionally, whereas pdpD (outer membrane protein in F. novicida; Ludu et al., 2008) is present in both Type A F. tularensis strains and F. novicida, pdpD has major deletions in Type B strains that likely result in two, small non-functional proteins (Bröms et al., 2010).
Secreted proteins
Various proteins have been reported to be secreted/released by F. tularensis but the mechanisms for their secretion are not known. Secretion could occur through one of the secretion systems outlined above, through outer membrane vesicle blebbing, or through yet to be elucidated mechanisms. Some studies have compared the secreted proteome of virulent and attenuated F. tularensis strains as a means to identify potential virulence factors. In one study, comparison of culture filtrate proteins from LVS and Schu S4 revealed three proteins detected in higher abundance from Schu S4, including the acid phosphatase AcpA, a beta-lactamase, and hypothetical protein FTT_0484 (Konecna et al., 2010). In another study, culture filtrates from a recent type A clinical isolate and LVS were compared, revealing 12 proteins likely released from both F. tularensis strains, including catalase-peroxidase KatG, chaperones DnaK, GroEL, and GroES, superoxide dismutase SodB, and bacterioferritin Bfr. Of these, KatG and DnaK were more prevalent from type A strain culture filtrates, Bfr was detected in multiple isoforms from LVS filtrates, and GroEL was released at equal levels from both F. tularensis strains. Interestingly, KatG and GroEL were detected in the cytosol of infected THP-1 cells, indicating that both may be important for intracellular replication (Lee et al., 2006).
AcpA has been found to be either associated with the outer membrane or secreted into culture supernatants by both F. novicida and LVS (Mohapatra et al., 2007; Dai et al., 2012). Importantly, AcpA also was isolated from the cytosol of infected macrophages, suggesting a role in virulence (Konecna et al., 2010; Dai et al., 2012). In addition to acid phosphatase activity, multiple functions have been attributed to F. tularensis AcpA, including phospholipase activity, lipase activity, ability to inhibit the neutrophil respiratory burst, general roles in intracellular growth, phagosomal escape, and virulence in animals (via i.n. or i.p. infection routes; Reilly et al., 1996; Mohapatra et al., 2007, 2008, 2010; McRae et al., 2010).
KatG has been noted to be important for in vitro detoxification of reactive oxygen and nitrogen species in both LVS and Schu S4 (Lindgren et al., 2007). As noted above, since Type A strains were found to express more KatG than LVS (Lee et al., 2006), it is interesting that LVS was found to be more susceptible to reactive oxygen and nitrogen species than virulent Type A or B strains (Lindgren et al., 2007). Despite the hypothetical role of KatG to protect F. tularensis from free radical damage, KatG was not found to be necessary for intracellular survival in macrophages for either LVS or Schu S4, but was required for LVS virulence in mice via the i.d. route (Lindgren et al., 2007).
Although far fewer studies have been performed to understand F. tularensis SodB, it has been found to confer in vitro bacterial resistance to peroxide and the superoxide generating compound paraquat. In addition, SodB was shown to be required for LVS virulence in mice by the i.n. route (Bakshi et al., 2006). The authors of that study proposed multiple modes of activity for SodB, including binding of free iron to limit highly lethal hydroxyl radical production, detoxification of superoxide to limit DNA, protein, and lipid toxicity, and detoxification of superoxide to prevent peroxynitrite formation (Bakshi et al., 2006).
Taken together, TolC and the FPI-encoded type VI-like secretion system appear to be the best candidates for F. tularensis secretion systems (Figure 1). Although many proteins have been reported to be released by F. tularensis (Table 3), there are major gaps in our understanding of F. tularensis secretion mechanisms and secretion machinery. Whereas studies on the F. novicida type VI-like secretion system are intriguing and have helped characterize the functions of various F. novicida FPI genes (Table 3), parallel studies in virulent F. tularensis strains are severely lacking, likely due to duplicate copies of the FPI in Type A and Type B strains. Future studies are needed to better understand F. tularensis secretion systems, secreted proteins, the role of the FPI in secretion, and confirm the existence of the Type VI secretion system in virulent F. tularensis strains.
Table 3.
Francisella secretion systems or proteins reported to be secreted.
| Gene name (if designated) | Schu S4 locus | LVS locus | F. novicida strain U112 locus | Proposed role |
|---|---|---|---|---|
| sodB | FTT0068 | FTL1791 | FTN1642 | Secreted |
| acpA | FTT0221 | FTL0158 | FTN0090 | Secreted |
| FTT0484 | FTL1583 | FTN0571 | Secreted | |
| β-lactamase | FTT0611c | FTL0879 | FTN1072 | Secreted |
| katG | FTT0721c | FTL1504 | FTN0633 | Secreted |
| ftlC | FTT1095c | FTL1107 | FTN0779 | Type I secretion |
| silC | FTT1258 | FTL0686 | FTN1277 | Type I secretion |
| dnaK | FTT1269c | FTL1191 | FTN1284 | Secreted |
| pdpA | FTT1344, FTT1699 |
FTL1172, FTL0126 |
FTN1309 | Type VI secretion |
| pdpB | FTT1345, FTT1700 |
FTL0125, FTL1171 |
FTN1310 | Type VI secretion |
| iglE | FTT1346, FTT1701 |
FTL0124, FTL1170 |
FTN1311 | Type VI secretion |
| vgrG | FTT1347, FTT1702 |
FTL0123, FTL1169 |
FTN1312 | Type VI secretion |
| iglF | FTT1348, FTT1703 |
FTL0122, FTL1168 |
FTN1313 | Type VI secretion |
| iglG | FTT1349, FTT1704 |
FTL0121, FTL1167 |
FTN1314 | Type VI secretion |
| iglH | FTT1350, FTT1705 |
FTL0120, FTL1166 |
FTN1315 | Type VI secretion |
| dotU | FTT1351, FTT1706 |
FTL0119, FTL1165 |
FTN1316 | Type VI secretion |
| iglI | FTT1352, FTT1707 |
FTL0118, FTL1164 |
FTN1317 | Type VI secretion |
| iglJ | FTT1353, FTT1708 |
FTL0117, FTL1163 |
FTN1318 | Type VI secretion |
| pdpC | FTT1354, FTT1709 |
FTL0116, FTL1162 |
FTN1319 | Type VI secretion |
| pdpE | FTT1355, FTT1710 |
FTL0115, FTL1161 |
FTN1320 | Type VI secretion |
| iglD | FTT1356c, FTT1711c |
FTL0114, FTL1160 |
FTN1321 | Type VI secretion |
| iglC | FTT1357c, FTT1712c |
FTL0113, FTL1159 |
FTN1322 | Type VI secretion |
| iglB | FTT1358c, FTT1713c |
FTL0112, FTL1158 |
FTN1323 | Type VI secretion |
| iglA | FTT1359c, FTT1714c |
FTL0111, FTL1157 |
FTN1324 | Type VI secretion |
| pdpD | FTT1360c, FTT1715c |
FTL0109, FTL0110, FTL1155, FTL1156 |
FTN1325 | Type VI secretion |
| anmK | FTT1361c, FTT1716c |
Absent | FTN1326 | Type VI secretion |
| bfr | FTT1441 | FTL0617 | FTN1410 | Secreted |
| groES | FTT1695 | FTL1715 | FTN1539 | Secreted |
| groEL | FTT1696 | FTL1714 | FTN1538 | Secreted |
| tolC | FTT1724c | FTL1865 | FTN1703 | Type I secretion |
Outer membrane proteins
Because of their surface localization, bacterial outer membrane proteins (OMPs) can play dual roles as virulence factors and as immune targets (Huntley et al., 2008; Confer and Ayalew, 2013; Embers and Narasimhan, 2013; Pore and Chakrabarti, 2013; Cash, 2014; Christodoulides, 2014). For these reasons, many investigators have attempted large-scale proteomic efforts to identify F. tularensis OMPs, including the use of biotinylation (Chandler et al., 2015), detergents (Twine et al., 2005b; Dresler et al., 2011), or sodium carbonate (Pavkova et al., 2005, 2006; Janovská et al., 2007) to extract and purify surface proteins. However, in each of those studies, large numbers of cytosolic, periplasmic, or proteins lacking transmembrane domains were identified, questioning the utility of those approaches. Despite these limitations, the following proteins were identified in three or more studies and are bioinformatically-predicted to be OMPs: FTT0077, FTT0087, FTT0188, FTT0209c, FTT0472, FTT0583 (FopA), FTT0721c (KatG), FTT0726c, FTT0842 (Pal), FTT0901 (Tul4-A), FTT1043 (Mip), FTT1103 (DsbA), FTT1441, FTT1483c, FTT1484c, FTT1572c, FTT1591, FTT1676, and FTT1747. To our knowledge, there have been few follow-up studies on these 19 proteins, so little is known about their true subcellular localization or their individual roles in virulence.
Gram-negative bacteria are known to spontaneously release outer membrane vesicles (OMVs) that are highly enriched in OMPs (Bonnington and Kuehn, 2014), thus it is not surprising that OMVs have been collected from F. novicida culture supernatants as a means to identify Francisella OMPs (Pierson et al., 2011; McCaig et al., 2013). However, mass spectrometry analysis of F. novicida OMVs indicated that OMPs compromised only 5–15% of identified proteins. Importantly, spontaneously-released OMVs have not been isolated from virulent F. tularensis strains, indicating that OMV shedding may be F. novicida-specific. Our laboratory developed and published a rigorous method to generate F. tularensis OMVs through spheroplasting and osmotic lysis, followed by OMV isolation using sucrose density gradient centrifugation (Huntley et al., 2007, 2010). This powerful technique resulted in the identification of 17 bona fide F. tularensis OMPs (Huntley et al., 2007) and confirmed some of the putative OMPs identified from large-scale proteomic efforts described above (e.g., FopA, KatG, Pal, Tul4-A, Mip, DsbA; Table 4). Additionally, our laboratory has demonstrated the protective potential of F. tularensis OMPs to serve as vaccines against Schu S4 challenge (Huntley et al., 2008).
Table 4.
F. tularensis outer membrane proteins (OMPs).
| Gene name (if designated) | Schu S4 locus | LVS locus | F. novicida strain U112 locus | Required for virulence? |
|---|---|---|---|---|
| fslE, srfA | FTT0025c | FTL1863 | FTN1686 | Yes |
| fsaP | FTT0119 | FTL1658 | FTN1596 | Yes |
| dipA | FTT0369c | FTL1306 | FTN0275 | Yes |
| fopA | FTT0583 | FTL1328 | FTN0756 | Yes |
| katG | FTT0721c | FTL1504 | FTN0633 | |
| FTT0825c | FTL0317 | FTN0340 | ||
| ompA | FTT0831c | FTL0325 | FTN0346 | Yes |
| pal | FTT0842 | FTL0336 | FTN0357 | |
| tul4-A | FTT0901 | FTL0421 | FTN0427 | |
| FTT0903 | FTL0423 | FTN0429 | ||
| tul4-B | FTT0904 | FTL0424 | FTN0430 | |
| yapH-N, fupA, fopC | FTT0918 | FTL0439 | FTN0444 | Yes |
| yapH-C, fupB | FTT0919 | FTL0439 | FTN0445 | |
| mip | FTT1043 | FTL1042 | FTN0921 | |
| tolC-A, ftlC | FTT1095c | FTL1107 | FTN0779 | |
| dsbA, fipB | FTT1103 | FTL1096 | FTN0771 | Yes |
| pilQ | FTT1156c | FTL0800 | FTN1137 | |
| SilC | FTT1258 | FTL0686 | FTN1277 | |
| iglE | FTT1346, FTT1701 |
FTL0124, FTL1170 |
FTN1311 | Yes |
| flpp3 | FTT1416c | FTL0645 | FTN1382 | Yes |
| ftaG | FTT1573c | FTL0535 | FTN1482 | |
| vacJ | FTT1591 | FTL1637 | FTN0322 | |
| iglE | FTT1701 | FTL1865 | FTN1703 | |
| tolC-B, tolC | FTT1724c | FTL0105 | FTN1734 | Yes |
| FTT1778c | FTL1863 | FTN1686 |
Aside from the above noted OMPs, the remainder of this section is devoted to OMPs that have been studied as F. tularensis virulence factors. FTT1416c first was identified as a F. tularensis membrane protein (Twine et al., 2005b). The LVS homolog, FTL0645, was identified from a transposon library as being required for LVS survival in the mouse lung (Su et al., 2007). Later analysis indicated that FTL0645 (named Flpp3) was a lipoprotein, stimulated TLR2, and reacted with antisera from LVS vaccinees and tularemia patients (Parra et al., 2010). The NMR structure of the Flpp3 Schu S4 homolog, FTT1416c, recently was solved and found to be structurally similar to the Bet v1 family of allergens, suggesting that FTT1416c could be a vaccine candidate (Zook et al., 2015). FTT1416c also was found to contain an internal hydrophobic cavity, suggesting that FTT1416c could be targeted for small molecule inhibitor studies (Zook et al., 2015).
Global transcriptional profiling of Schu S4 from mouse macrophages revealed that FTT0369c (DipA) was upregulated 2- to 3-fold and a ΔFTT0369c mutant confirmed that FTT0369c was required for virulence both in vitro and in vivo (i.n. and i.d. routes tested in mice; Wehrly et al., 2009). More comprehensive studies demonstrated that Schu S4 ΔdipA mutants escaped from phagosomes, failed to replicate in the cytosol, and then were targeted to autophagy vacuoles (LAMP1+, LC3+) for clearance (Chong et al., 2012). Biochemical and structural analysis of DipA showed the protein to be outer membrane-localized and surface-exposed in SchuS4. In addition, DipA was found to interact with OMP FopA, suggesting that DipA and FopA form an outer membrane complex (Chong et al., 2013). A more recent study identified DipA as a DsbA substrate and demonstrated that, although DipA contains a putative lipoprotein acylation motif, DipA is not lipidated (Ren et al., 2014).
An OmpA-like protein, FTT0831c in Schu S4 and FTL0325 in LVS, was found to be surface-exposed in LVS and outer membrane-localized in Schu S4 (Mahawar et al., 2012). FTL0325 and FTT0831c are required for macrophage replication and deletion of the gene in each respective strain caused increased proinflammatory cytokine production in macrophages (Mahawar et al., 2012). Ectopic expression of FTT0831c in a mammalian cell line blocked NFκB translocation to the nucleus and suggested that FTL0325/FTT0831c functions to suppress proinflammatory cytokine responses in host cells (Mahawar et al., 2012). The LVS FTL0325 mutant was attenuated in an i.n. mouse infection model, yet still induced high levels of inflammatory cytokines from days one through four post-infection, as compared to wild-type LVS or other LVS mutants (Mahawar et al., 2013). These early pro-inflammatory cytokine responses to FTL0325 were found to correlate with protection against Schu S4 challenge. A later study noted that FTT0831c/FTL0325 mutants had reduced viability in laboratory medium, resulted in small colony phenotypes on agar, were larger and more spherical than wild-type cells, and had an irregular morphology with blebs near the bacterial midpoint and enlarged bacterial tips. Based on these abnormalities, the authors proposed that FTT0831c/FTL0325 acts as a structural protein to link the F. tularensis outer membrane to peptidoglycan (Robertson et al., 2014). The authors' results suggested that loss of FTT0831c/FTL0325 resulted in altered cell morphology and decreased viability, likely resulting in increased release or exposure of PAMPs, which may have explained the increased proinflammatory cytokine response previously observed.
FTT0918 first was characterized as a 58-kDa protein required for Schu S4 virulence in vitro and in vivo (i.d. mice infections; Twine et al., 2005a). Interestingly, in LVS, the FTT0918 homolog is C-terminally truncated and fused in-frame with the N-terminally truncated downstream FTT0919 homolog, resulting in a hybrid protein FTL0439 (Twine et al., 2005a; Huntley et al., 2007). Despite coding differences between Schu S4 and LVS, FTT0918 and FTL0439 both are outer membrane-localized (Huntley et al., 2007). The importance of FTT0918 in F. tularensis virulence was further highlighted by a study that reintroduced PilA and FTT0918 into LVS, which restored virulence to wild-type Type B levels (Salomonsson et al., 2009). The F. novicida homolog, FTN0444 (named FopC), appears to play similar roles in virulence both in vitro and in vivo (i.n. mice infections; Nallaparaju et al., 2011). In vitro studies demonstrated that, as compared to wild-type F. novicida, F. novicida FopC mutants were more susceptible to IFNγ-induced macrophage killing and had decreased membrane stability (Nallaparaju et al., 2011). FTT0918 also has been shown to be important for both siderophore-dependent and siderophore-independent (ferrous) iron acquisition mechanisms in Schu S4 (Lindgren et al., 2009; Ramakrishnan et al., 2012). In this context, FTT0918 has been referred to as FupA, for Fe utilization protein A (Lindgren et al., 2009). Interestingly, despite its role in iron uptake, fupA expression is not regulated by iron limitation (Lindgren et al., 2009; Ramakrishnan et al., 2012). It is well-known that iron is critical for many intracellular pathogens (Cassat and Skaar, 2013; Caza and Kronstad, 2013), thus it is not surprising that FupA-mediated iron uptake was found to be important for Schu S4 virulence both in vitro and in vivo (sub-q and i.d. routes tested in mice; Lindgren et al., 2009; Ramakrishnan et al., 2012). In LVS, the homologous hybrid protein, FupA/B, has been shown to play similar roles in uptake of both ferrous and siderophore-bound iron (Sen et al., 2010; Ramakrishnan and Sen, 2014). Similarly, FupA/B was found to be important for intracellular growth and for LVS virulence in i.p. infected mice (Sen et al., 2010).
FslE (FTT0025c) is a second F. tularensis OMP (Huntley et al., 2007) that has been shown to play a role in iron uptake (Ramakrishnan et al., 2008, 2012; Ramakrishnan and Sen, 2014). In Schu S4, FslE appears to be primarily responsible for siderophore-mediated ferric iron acquisition (Ramakrishnan et al., 2008, 2012). Schu S4 FslE was found to be essential for growth in iron-limiting conditions and, as compared to FupA (FTT0918), FslE expression was increased during growth in iron-limitation (Ramakrishnan et al., 2008, 2012). By comparison, LVS FslE appears to play a secondary role in siderophore-mediatated iron uptake, as FslE was not required for the growth of LVS in iron-limiting conditions (Sen et al., 2010). Interestingly, a Schu S4 fslE mutant was not attenuated for intracellular growth or virulence in sub-q-infected mice, but a fupA–fslE double mutant was more severely attenuated than a fupA mutant in sub-q infected mice, suggesting that FslE and FupA play synergistic roles in vivo (Ramakrishnan et al., 2012). Similarly, LVS fslE mutants were not attenuated in i.p.-infected mice, but a fslE-fupA/B double mutant was more severely attenuated than a fupA/B single mutant in i.p.-infected mice, further supporting the synergistic roles of FslE and FupA/B in F. tularensis virulence (Sen et al., 2010).
DsbA (FTT1103) was identified as a putative virulence factor through a Schu S4 transposon library screen that identified mutants defective in intracellular growth (Qin and Mann, 2006). A subsequent full-gene deletion confirmed that FTT1103 was required for intracellular survival, phagosomal escape, and virulence in mice (i.n. and sub-q routes tested; Qin et al., 2009). A number of follow-up studies have implicated DsbA (FTT1103 in Schu S4, FTL1096 in LVS, also referred to as FipB) as an essential F. tularensis virulence factor (Qin and Mann, 2006; Thakran et al., 2008; Qin et al., 2009, 2011, 2014; Straskova et al., 2009; Schmidt et al., 2013; Senitkova et al., 2015). However, given the well-known function of bacterial DsbA proteins to form disulfide bonds in various substrates, including true virulence factors, it seems unlikely that F. tularensis DsbA directly promotes host cell binding, invasion, intracellular survival, or other virulence functions. Further, F. tularensis DsbA is unusual in that it is outer membrane-localized and lipidated, as compared to the periplasmically-localized DsbA in other bacteria (Huntley et al., 2007; Thakran et al., 2008). F. tularensis DsbA lipidation has been demonstrated to stimulate the TLR2/TLR1 heterodimer and induce proinflammatory chemokine expression (Thakran et al., 2008). Additionally, F. tularensis DsbA has been reported to be glycosylated, although the role of this glycosylation is unknown (Balonova et al., 2012). Finally, F. tularensis DsbA is distinct from other bacterial DsbA orthologs in that it contains an N-terminal Forskolin-binding protein-N (FKBP-N) dimerization domain as well as a C-terminal DsbA-like domain (Qin et al., 2011). The function of the DsbA N-terminal FKBP-N domain is somewhat controversial, as this domain has been reported to possess isomerase and chaperone activities, was reported to be required for DsbA dimerization, and was shown to be required for virulence in mice by either i.n. or sub-q infection routes (Qin et al., 2014; Senitkova et al., 2015). Conversely, other studies have reported that the FKBP-N domain does not possess chaperone activity, is not required for dimerization, and is not required for virulence in mice by either i.n. or sub-q infection routes (Schmidt et al., 2013; Qin et al., 2014). More thorough analyses are needed to clarify the true function of the DsbA FKBP-N domain. The C-terminal DsbA-like domain has been more carefully studied, consistently demonstrating that the thioredoxin-like CXXC active site motif is required for in vitro and mouse virulence (i.n. and sub-q routes tested; Qin et al., 2011; Schmidt et al., 2013). The DsbA-like domain also has been shown to possess disulfide oxidoreductase activity (Straskova et al., 2009; Schmidt et al., 2013; Qin et al., 2014). Work by our laboratory recently revealed that F. tularensis DsbA is a bi-functional protein, possessing both oxidoreductase and isomerase activities to correctly form complex disulfide bonds in over 50 substrates, including some of the above noted virulence factors (e.g., DipA, FopA, and FTT0831c), type IV pilus components, pathogenicity island proteins, previously described OMPs, and 25 hypothetical proteins (many of which are predicted OMPs). As proof-of-principal that these newly-identified DsbA substrates were true virulence factors, we demonstrated that at least two novel proteins, FTL1548 and FTL1709, were required for LVS i.n. infection of mice (Ren et al., 2014).
Among other F. tularensis OMPs that have been studied, FsaP (FTL1658) was shown to be a surface-exposed OMP important for LVS adherence to epithelial cells (Melillo et al., 2006). Overexpression of FsaP in E. coli increased bacterial adherence to epithelial cells, confirming the LVS findings. Interestingly, despite 98% sequence identity to its LVS and Schu S4 (FTT0119) homologs, the F. novicida FsaP (FTN1596) homolog has an amino acid substitution in its signal sequence that appears to affect FsaP localization and function, as F. novicida FsaP was not outer membrane-localized and F. novicida did not adhere to epithelial cells as well as LVS (Melillo et al., 2006). Already highlighted in the Secretion Systems and Secreted Proteins Section above are two FPI members and reported components of a Type VI-like secretion system, outer membrane lipoprotein IglE (FTT1346/FTT1701) and putative outer membrane protein PdpD (outer membrane localization only shown in F. novicida; Ludu et al., 2008). Also highlighted in the Secretion Systems and Secreted Proteins Section were OMPs and TolC homologs TolC (FTT1724c), FtlC (FTT1095c), and SilC (FTT1258). Of these, only TolC has been shown to be involved in virulence.
In summary, the large number of reported F. tularensis OMPs and studies to understand their function (Table 4) highlight the importance of OMPs as potential virulence factors and vaccine targets (Figure 1). Future OMP identification and characterization studies will help to better understand the extreme virulence of F. tularensis.
Periplasmic and inner membrane proteins
In Gram-negative bacteria, the periplasm and inner membrane (IM) compartmentalize and protect the cytoplasmic contents from the harsh external environment. In E. coli, more than 300 periplasmic proteins perform a variety of functions, including folding proteins into their native confirmation, sensing unfolded or damaged proteins (chaperones), degrading proteins, or binding/transporting sugars, amino acids, and other nutrients between the OM and IM (Merdanovic et al., 2011; Goemans et al., 2014). The IM is vitally important for many processes, including energy production, lipid biosynthesis, transport of sugars, amino acids, and other nutrients into the cytoplasm, secretion of proteins out of the cytoplasm, and signal transduction (Silhavy et al., 2010; Luirink et al., 2012). Despite the essential nature of these proteins for bacterial viability, only a few F. tularensis periplasmic or IM proteins have been studied and correlated with virulence.
SodC (FTL0380) is predicted to be a periplasmic protein related to the copper/zinc-containing superoxide dismutase family. LVS SodC was found to be important for peroxide and superoxide resistance in vitro, but was not involved in reactive nitrogen resistance. In IFN-γ-activated macrophages, sodC mutants were attenuated for macrophage growth and were found to be partially attenuated in a mouse i.n. infection model (Melillo et al., 2009).
RipA (FTL1914), an IM protein, first was identified by screening a LVS transposon library for mutants that failed to replicate in alveolar epithelial cells (Fuller et al., 2008). Although RipA is conserved among all sequenced Francisella strains, it has few orthologs in other bacteria, and its function is not completely understood. In vitro analyses demonstrated that the ΔripA mutant invaded macrophages and epithelial cells as efficiently as wild-type LVS, but the ΔripA mutant had significant survival defects in both cell types. More specifically, the ΔripA mutant escaped from phagosomes and re-entered autophagy-like vacuoles as efficiently as wild-type LVS, but was unable to survive in the host cell cytoplasm. In vivo, the ΔripA mutant failed to replicate in mouse lungs following i.n. challenge and was severely attenuated for dissemination to livers and spleens of infected mice (Fuller et al., 2008). pH appears to play a role in the ΔripA cytosolic replication defect, as the ΔripA mutant replicated similar to wild-type LVS in media at pH 6.2, but the ΔripA mutant replicated poorly at pH 7.5 (similar to the pH in the macrophage cytosol). Further, ripA transcription was found to be upregulated at pH 7.5, compared with gene expression at pH 5.5 (Fuller et al., 2009). Two studies noted that RipA plays a role in immune evasion, as the ΔripA mutant induced inflammasome activation, including IL-1β, IL-18, and TNF-α, and cell death in macrophages (Huang et al., 2010; Mortensen et al., 2012). However, in F. novicida, a ΔripA mutant was found to possess aberrant cell morphology and lysed more readily inside macrophages than wild-type F. novicida, suggesting that ΔripA mutants induced inflammasome activation due to PAMP release (Peng et al., 2011). A separate study showed that RipA interacts with and modulates LpxA, a component of the Lipid A biosynthesis pathway (Miller et al., 2014). Taken together, it appears that RipA may not actively suppress immune responses, but instead may modulate lipid A/LPS expression inside host cells and promote intracellular survival (Miller et al., 2014).
The KdpDE two component signaling system has been shown to be important for potassium ion homeostasis and virulence in diverse bacterial species (Freeman et al., 2013). All F. tularensis strains encode the IM-bound sensor kinase KdpD (FTT1736c, FTL1879, FTN1715). However, the tandemly-arranged response regulator KdpE (FTN1714) only is present in F. novicida and not in virulent Francisella strains. F. novicida KdpD was shown to be the primary kinase that phosphorylated the orphan response regulator PmrA (FTT1557c, FTL0552, FTN1465) and this phosphorylation increased PmrA binding to pmrA and pdpD promoters. Phosphorylation of PmrA also was found to be important for virulence in macrophages and i.n. infection of mice (Bell et al., 2010). Other studies have confirmed the role of F. novicida KpdD for in vivo virulence, including in Drosophila (Moule et al., 2010) and mice (sub-q and i.p. routes tested; Weiss et al., 2007) infection models. F. novicida ΔkdpD mutants also were found to be more sensitive to reactive oxygen species, but not antimicrobial peptides in vitro (Moule et al., 2010).
QseBC is a two-component signaling system found in many bacterial pathogens that responds to host stress hormones epinephrine and norepinephrine, as well as the bacterial signal AI-3 (Clarke et al., 2006; Curtis et al., 2014; Weigel and Demuth, 2015). In F. tularensis, the IM sensor kinase QseC (FTT0094c, FTL1762, FTN1617) is not transcriptionally linked to a response regulator, although homology searches suggest that PmrA (FTT1557c, FTL0552, FTN1465; described above) may be a QseB homolog (Durham-Colleran et al., 2010). F. novicida QseC mutants were found to be attenuated in Drosophila (Moule et al., 2010) and mice (i.p. and sub-q routes tested; Weiss et al., 2007) infection models. In LVS, a ΔqseC mutant was extremely serum sensitive, could no longer synthesize high molecular mass O-polysaccharide in its LPS, and was attenuated for macrophage growth and virulence in mice via the sub-q route (Mokrievich et al., 2010). QseC also has been reported to be important for F. novicida biofilm production (Durham-Colleran et al., 2010). In a separate study, maprotiline, an FDA-approved norepinephrine uptake inhibitor, was found to inhibit F. novicida biofilm production in a QseC dependent manner and prolonged survival in a mouse i.n. infection model (Dean and van Hoek, 2015). Due to the broad distribution of QseC across many pathogenic bacteria, QseC is an attractive target for anti-virulence drug development efforts (Rasko et al., 2008; Curtis et al., 2014; Dean and van Hoek, 2015). Indeed, Schu S4 QseC was found to be upregulated during mammalian infection and a small-molecule inhibitor of QseC, LED209, decreased expression of many F. tularensis virulence genes and attenuated intracellular survival and virulence in mice in an i.n. infection model (Rasko et al., 2008; Curtis et al., 2014).
Four other IM proteins, LpxE (Wang et al., 2004), LpxF (Wang et al., 2006, 2007), KdoH1, and KdoH2 (Zhao and Raetz, 2010), have been implicated in F. tularensis virulence and antimicrobial resistance, but their roles in LPS biosynthesis and modification suggest an indirect role. Finally, although SecY, LolC, and Cht1 were shown to be IM-localized in Schu S4 and LVS (Huntley et al., 2007, 2010) their roles in virulence are not known.
In summary, given the fairly limited number of studies performed to identify F. tularensis periplasmic and IM proteins and examine their roles in virulence (Table 5), more emphasis should be devoted to studying these potentially-important proteins. As noted in the beginning of this section, given the large number of periplasmic proteins in other Gram-negative bacteria and critical role of IM proteins in bacterial survival, periplasmic and IM proteins may be an understudied area with great potential to reveal new F. tularensis virulence mechanisms.
Table 5.
F. tularensis periplasmic and inner membrane proteins (IMPs).
| Gene name (if designated) | Schu S4 locus | LVS locus | F. novicida strain U112 locus | Bacterial location |
|---|---|---|---|---|
| qseC | FTT0094c | FTL1762 | FTN1617 | Inner membrane |
| ripA | FTT0181c | FTL1914 | FTN0157 | Inner membrane |
| secY | FTT0345 | FTL0256 | FTN0259 | Inner membrane |
| kdoH2 | FTT0398c | FTL0464 | FTN0494 | Inner membrane |
| kdoH1 | FTT0399c | FTL0465 | FTN0495 | Inner membrane |
| lolC | FTT0404 | FTL0474 | FTN0502 | Inner membrane |
| cht1 | FTT0715 | FTL1521 | FTN0627 | Inner membrane |
| sodC | FTT0879 | FTL0380 | FTN0405 | Periplasm |
| lpxE | FTT0891 | FTL0393 | FTN0416 | Inner membrane |
| lpxF | FTT1643c | FTL1815 | FTN0286 | Inner membrane |
| kdpD | FTT1736c | FTL1879 | FTN1715 | Inner membrane |
Summary and perspectives
Since 2001, there has been a surge in research studies conducted on F. tularensis and its non-pathogenic relative F. novicida. Whereas, these studies have provided a wealth of information about many previously unstudied F. tularensis hypothetical proteins and have revealed novel virulence strategies, major questions still remain about the true role of many of these putative virulence factors in Type A and Type B virulent strains. In this review, we focused on components of the F. tularensis envelope that have been reported to contribute to immune evasion, intracellular replication and survival, and mammalian virulence. Since the envelope is the outermost layer of F. tularensis, many studies have aimed to identify and characterize envelope components, given their potential surface exposure and likely direct interaction with host cell molecules. First, the F. tularensis capsule has been shown to be identical to the O-antigen, blocks both IgM and complement component C3 binding, and is required for in vitro and in vivo virulence of many F. tularensis strains, including Schu S4 (Figure 1). However, capsule expression appears to be variable and overlap between capsule and LPS synthesis pathways (Table 1) brings into question if F. tularensis capsule is real or if the bacterium simply expresses free O-antigen that aggregates near the surface. These questions highlight the need for future studies to better understand capsule. Second, F. tularensis has evolved a unique LPS with many structural variations from typical Gram-negative LPS, including fewer acyl chains (tetraacylated), longer fatty acid chains (16–20 carbons), and reduced or no lipid A phosphorylation (Figure 1). Importantly, F. tularensis also is able to subtly alter expression or components of its LPS, including O-antigen and lipid A, which affect outer membrane stability and immunogenicity. Together, these LPS alterations affect animal virulence, immune activation/evasion, and the enigmatic blue-gray colony phenotype that plagues the field. As noted above, since F. tularensis capsule and LPS appear to share some biosynthetic pathways (Table 1) and serum resistance has been linked to both capsule and LPS, future studies are needed to resolve how these envelope components, either separately or cooperatively, contribute to serum resistance. Third, type IV pili are responsible for thin fibers or horn-like protrusions observed on the surface of many F. tularensis strains and have been shown to be required for F. tularensis virulence (Figure 1). Although there has been some speculation that components of F. novicida type IV pili may be involved in a putative type II-like secretion system, these preliminary findings have not been confirmed in virulent F. tularensis strains. Interestingly, despite the well-known role of type IV pili in adhesion, aggregation, twitching motility, and DNA uptake in other bacteria, similar functions have not been attributed to F. tularensis type IV pili. Whereas, a number of studies have demonstrated that F. tularensis type IV pili are important for Type A and Type B virulence, more detailed studies are needed to elucidate the mechanisms by which type IV pili interact with or damage the host. Fourth, a number of investigators have examined culture filtrate proteins from various F. tularensis strains and identified proteins that presumably are secreted (Table 3). Indeed, whereas there is much interest in identifying F. tularensis secretion systems and secreted proteins to help explain how the bacterium blocks phagosome-lysosome fusion or escapes from the phagosome, there still is a dearth of information about F. tularensis secretion. One possibility is that the F. tularensis TolC orthologs, including TolC, FtlC, and SilC (Table 3), function as a Type I secretion system to release effector molecules (Figure 1). Of these, TolC has been shown to play a role in Schu S4 and LVS virulence. Another possibility is that the F. novicida FPI-encoded Type VI-like secretion system delivers effector proteins to host cells (Figure 1). However, the existence of a Type VI-like secretion system in virulent F. tularensis has not been reported. Fifth, at least 10 OMPs have been shown to be required for F. tularensis virulence (Table 4 and Figure 1). Although OMP functions are diverse, these studies highlight the importance of identifying and characterizing OMP function to better understand F. tularensis virulence. Sixth, although few studies exist on F. tularensis periplasmic or inner membrane proteins, the existing studies indicate that the periplasm plays a key role in detoxifying free radicals and the inner membrane contains two component signaling systems important for altering gene expression during infection (Figure 1).
In conclusion, the F. tularensis envelope is a dynamic and robust outer structure that protects the bacterium from immune recognition and clearance, facilitates host cell invasion and survival, and promotes infection and disease. Despite many advances over the decades and in recent years, much more work is needed to better understand the broad host range, environmental persistence, and extreme virulence of this complex pathogen.
Author contributions
HR and JH reviewed all listed references, compiled information for all tables and figures, and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
HR and JH are supported by grant CBCall14-CBM-02-1-0024 from the Defense Threat Reduction Agency (DTRA), U.S. Department of Defense (DoD). JH is additionally supported by grant R01 AI093351 from the National Institute of Allergy and Infectious Disease of the National Institutes of Health (NIAID-NIH). This content is solely the responsibility of the authors and does not necessarily represent the official views of the Department of Defense or the National Institutes of Health.
References
- Ancuta P., Pedron T., Girard R., Sandström G., Chaby R. (1996). Inability of the Francisella tularensis lipopolysaccharide to mimic or to antagonize the induction of cell activation by endotoxins. Infect. Immun. 64, 2041–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apicella M. A., Post D. M., Fowler A. C., Jones B. D., Rasmussen J. A., Hunt J. R., et al. (2010). Identification, characterization and immunogenicity of an O-antigen capsular polysaccharide of Francisella tularensis. PLoS ONE 5:e11060. 10.1371/journal.pone.0011060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ark N. M., Mann B. J. (2011). Impact of Francisella tularensis pilin homologs on pilus formation and virulence. Microb. Pathog. 51, 110–120. 10.1016/j.micpath.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi C. S., Malik M., Regan K., Melendez J. A., Metzger D. W., Pavlov V. M., et al. (2006). Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 188, 6443–6448. 10.1128/JB.00266-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balonova L., Mann B. F., Cerveny L., Alley W. R., Jr., Chovancova E., Forslund A. L., et al. (2012). Characterization of protein glycosylation in Francisella tularensis subsp. holarctica: identification of a novel glycosylated lipoprotein required for virulence. Mol. Cell. Proteomics 11:M111.015016. 10.1074/mcp.M111.015016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara A. B., Champion A. E., Wang X., Berg G., Apicella M. A., McLendon M., et al. (2011). Isolation and mutagenesis of a capsule-like complex (CLC) from Francisella tularensis, and contribution of the CLC to F. tularensis virulence in mice. PLoS ONE 6:e19003. 10.1371/journal.pone.0019003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barel M., Charbit A. (2013). Francisella tularensis intracellular survival: to eat or to die. Microbes Infect. 15, 989–997. 10.1016/j.micinf.2013.09.009 [DOI] [PubMed] [Google Scholar]
- Barel M., Ramond E., Gesbert G., Charbit A. (2015). The complex amino acid diet of Francisella in infected macrophages. Front. Cell. Infect. Microbiol. 5:9. 10.3389/fcimb.2015.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. H., Kaufman J. W., Zhang D. S., Weiss J. P. (2014). Metabolic labeling to characterize the overall composition of Francisella lipid A and LPS grown in broth and in human phagocytes. Innate Immun. 20, 88–103. 10.1177/1753425913485308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. H., Weiss J., Apicella M. A., Nauseef W. M. (2006). Basis for the failure of Francisella tularensis lipopolysaccharide to prime human polymorphonuclear leukocytes. Infect. Immun. 74, 3277–3284. 10.1128/IAI.02011-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. R., Chong A., Wehrly T. D., Yu J. J., Rodriguez S. A., Liu J., et al. (2009). The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74, 1459–1470. 10.1111/j.1365-2958.2009.06947.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley A. S., Cotter R. J., Vogel S. N., Inzana T. J., Qureshi A. A., Qureshi N. (2012). A variety of novel lipid A structures obtained from Francisella tularensis live vaccine strain. Innate Immun. 18, 268–278. 10.1177/1753425911401054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B. L., Mohapatra N. P., Gunn J. S. (2010). Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infect. Immun. 78, 2189–2198. 10.1128/IAI.00021-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J. L., Pelicic V. (2015). Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol. Rev. 39, 134–154. 10.1093/femsre/fuu001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnington K. E., Kuehn M. J. (2014). Protein selection and export via outer membrane vesicles. Biochim. Biophys. Acta 1843, 1612–1619. 10.1016/j.bbamcr.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Lavander M., Meyer L., Sjöstedt A. (2011). IglG and IglI of the Francisella pathogenicity island are important virulence determinants of Francisella tularensis LVS. Infect. Immun. 79, 3683–3696. 10.1128/IAI.01344-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Lavander M., Sjöstedt A. (2009). A conserved alpha-helix essential for a type VI secretion-like system of Francisella tularensis. J. Bacteriol. 191, 2431–2446. 10.1128/JB.01759-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Meyer L., Lavander M., Larsson P., Sjöstedt A. (2012a). DotU and VgrG, core components of type VI secretion systems, are essential for Francisella LVS pathogenicity. PLoS ONE 7:e34639. 10.1371/journal.pone.0034639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Meyer L., Sun K., Lavander M., Sjöstedt A. (2012b). Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS ONE 7:e50473. 10.1371/journal.pone.0050473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Sjöstedt A., Lavander M. (2010). The Role of the Francisella tularensis Pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front. Microbiol. 1:136. 10.3389/fmicb.2010.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkinshaw B. J., Strynadka N. C. (2014). Assembly and structure of the T3SS. Biochim. Biophys. Acta 1843, 1649–1663. 10.1016/j.bbamcr.2014.01.035 [DOI] [PubMed] [Google Scholar]
- Carvalho C. L., Lopes de Carvalho I., Zé-Zé L., Núncio M. S., Duarte E. L. (2014). Tularaemia: a challenging zoonosis. Comp. Immunol. Microbiol. Infect. Dis. 37, 85–96. 10.1016/j.cimid.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash P. (2014). Proteomic analysis of uropathogenic Escherichia coli. Expert Rev. Proteomics 11, 43–58. 10.1586/14789450.2014.877845 [DOI] [PubMed] [Google Scholar]
- Cassat J. E., Skaar E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13, 509–519. 10.1016/j.chom.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza M., Kronstad J. W. (2013). Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 3:80. 10.3389/fcimb.2013.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli J., Zahrt T. C. (2013). Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010314. 10.1101/cshperspect.a010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Monfett M., Maier T. M., Benach J. L., Frank D. W., Thanassi D. G. (2008). Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect. Immun. 76, 2852–2861. 10.1128/IAI.01726-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion M. D., Zeng Q., Nix E. B., Nano F. E., Keim P., Kodira C. D., et al. (2009). Comparative genomic characterization of Francisella tularensis strains belonging to low and high virulence subspecies. PLoS Pathog. 5:e1000459. 10.1371/journal.ppat.1000459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J. C., Sutherland M. D., Harton M. R., Molins C. R., Anderson R. V., Heaslip D. G., et al. (2015). Francisella tularensis LVS surface and membrane proteins as targets of effective post-exposure immunization for tularemia. J. Proteome Res. 14, 664–675. 10.1021/pr500628k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherwonogrodzky J. W., Knodel M. H., Spence M. R. (1994). Increased encapsulation and virulence of Francisella tularensis live vaccine strain (LVS) by subculturing on synthetic medium. Vaccine 12, 773–775. 10.1016/0264-410X(94)90284-4 [DOI] [PubMed] [Google Scholar]
- Chong A., Child R., Wehrly T. D., Rockx-Brouwer D., Qin A., Mann B. J., et al. (2013). Structure-function analysis of DipA, a virulence factor required for intracellular replication. PLoS ONE 8:e67965. 10.1371/journal.pone.0067965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A., Wehrly T. D., Child R., Hansen B., Hwang S., Virgin H. W., et al. (2012). Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy 8, 1342–1356. 10.4161/auto.20808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P. J., Whitaker N., González-Rivera C. (2014). Mechanism and structure of the bacterial type IV secretion systems. Biochim. Biophys. Acta 1843, 1578–1591. 10.1016/j.bbamcr.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulides M. (2014). Neisseria proteomics for antigen discovery and vaccine development. Expert Rev. Proteomics 11, 573–591. 10.1586/14789450.2014.938640 [DOI] [PubMed] [Google Scholar]
- Clarke M. B., Hughes D. T., Zhu C., Boedeker E. C., Sperandio V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 10420–10425. 10.1073/pnas.0604343103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay C. D., Soni S., Gunn J. S., Schlesinger L. S. (2008). Evasion of complement-mediated lysis and complement C3 deposition are regulated by Francisella tularensis lipopolysaccharide O antigen. J. Immunol. 181, 5568–5578. 10.4049/jimmunol.181.8.5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens D. L., Ge P., Lee B. Y., Horwitz M. A., Zhou Z. H. (2015). Atomic structure of T6SS reveals interlaced array essential to function. Cell 160, 940–951. 10.1016/j.cell.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens D. L., Lee B. Y., Horwitz M. A. (2012). O-antigen-deficient Francisella tularensis Live Vaccine Strain mutants are ingested via an aberrant form of looping phagocytosis and show altered kinetics of intracellular trafficking in human macrophages. Infect. Immun. 80, 952–967. 10.1128/IAI.05221-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confer A. W., Ayalew S. (2013). The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet. Microbiol. 163, 207–222. 10.1016/j.vetmic.2012.08.019 [DOI] [PubMed] [Google Scholar]
- Costa T. R., Felisberto-Rodrigues C., Meir A., Prevost M. S., Redzej A., Trokter M., et al. (2015). Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. 10.1038/nrmicro3456 [DOI] [PubMed] [Google Scholar]
- Cote C. K., Welkos S. L., Bozue J. (2011). Key aspects of the molecular and cellular basis of inhalational anthrax. Microbes Infect. 13, 1146–1155. 10.1016/j.micinf.2011.07.005 [DOI] [PubMed] [Google Scholar]
- Cowley S. C. (2009). Editorial: Proinflammatory cytokines in pneumonic tularemia: too much too late? J. Leukoc. Biol. 86, 469–470. 10.1189/jlb.0309119 [DOI] [PubMed] [Google Scholar]
- Cowley S. C., Elkins K. L. (2011). Immunity to Francisella. Front. Microbiol. 2:26. 10.3389/fmicb.2011.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley S. C., Myltseva S. V., Nano F. E. (1996). Phase variation in Francisella tularensis affecting intracellular growth, lipopolysaccharide antigenicity and nitric oxide production. Mol. Microbiol. 20, 867–874. 10.1111/j.1365-2958.1996.tb02524.x [DOI] [PubMed] [Google Scholar]
- Craig L., Pique M. E., Tainer J. A. (2004). Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2, 363–378. 10.1038/nrmicro885 [DOI] [PubMed] [Google Scholar]
- Curtis M. M., Russell R., Moreira C. G., Adebesin A. M., Wang C., Williams N. S., et al. (2014). QseC inhibitors as an antivirulence approach for Gram-negative pathogens. MBio 5:e02165. 10.1128/mBio.02165-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Mohapatra N. P., Schlesinger L. S., Gunn J. S. (2012). The acid phosphatase AcpA is secreted in vitro and in macrophages by Francisella spp. Infect. Immun. 80, 1088–1097. 10.1128/IAI.06245-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Rajaram M. V., Curry H. M., Leander R., Schlesinger L. S. (2013). Fine tuning inflammation at the front door: macrophage complement receptor 3-mediates phagocytosis and immune suppression for Francisella tularensis. PLoS Pathog. 9:e1003114. 10.1371/journal.ppat.1003114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean S. N., van Hoek M. L. (2015). Screen of FDA-approved drug library identifies maprotiline, an antibiofilm and antivirulence compound with QseC sensor-kinase dependent activity in Francisella novicida. Virulence 6, 487–503. 10.1080/21505594.2015.1046029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin O. M., Duplantis B. N., Ludu J. S., Hare R. F., Nix E. B., Schmerk C. L., et al. (2011). The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology (Reading Engl). 157, 3483–3491. 10.1099/mic.0.052308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin O. M., Ludu J. S., Nano F. E. (2007). The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 7:1. 10.1186/1471-2180-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis D. T., Inglesby T. V., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., et al. (2001). Tularemia as a biological weapon: medical and public health management. JAMA 285, 2763–2773. 10.1001/jama.285.21.2763 [DOI] [PubMed] [Google Scholar]
- Doyle C. R., Pan J. A., Mena P., Zong W. X., Thanassi D. G. (2014). TolC-dependent modulation of host cell death by the Francisella tularensis live vaccine strain. Infect. Immun. 82, 2068–2078. 10.1128/IAI.00044-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresler J., Klimentova J., Stulik J. (2011). Francisella tularensis membrane complexome by blue native/SDS-PAGE. J. Proteomics 75, 257–269. 10.1016/j.jprot.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Durham-Colleran M. W., Verhoeven A. B., van Hoek M. L. (2010). Francisella novicida forms in vitro biofilms mediated by an orphan response regulator. Microb. Ecol. 59, 457–465. 10.1007/s00248-009-9586-9 [DOI] [PubMed] [Google Scholar]
- Egge-Jacobsen W., Salomonsson E. N., Aas F. E., Forslund A. L., Winther-Larsen H. C., Maier J., et al. (2011). O-linked glycosylation of the PilA pilin protein of Francisella tularensis: identification of the endogenous protein-targeting oligosaccharyltransferase and characterization of the native oligosaccharide. J. Bacteriol. 193, 5487–5497. 10.1128/JB.00383-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigelsbach H. T., Braun W., Herring R. D. (1951). Studies on the variation of Bacterium tularense. J. Bacteriol. 61, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigelsbach H. T., Downs C. M. (1961). Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J. Immunol. 87, 415–425. [PubMed] [Google Scholar]
- Elkins K. L., Cowley S. C., Bosio C. M. (2007). Innate and adaptive immunity to Francisella. Ann. N. Y. Acad. Sci. 1105, 284–324. 10.1196/annals.1409.014 [DOI] [PubMed] [Google Scholar]
- Ellis J., Oyston P. C., Green M., Titball R. W. (2002). Tularemia. Clin. Microbiol. Rev. 15, 631–646. 10.1128/CMR.15.4.631-646.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embers M. E., Narasimhan S. (2013). Vaccination against Lyme disease: past, present, and future. Front. Cell. Infect. Microbiol. 3:6. 10.3389/fcimb.2013.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund A. L., Kuoppa K., Svensson K., Salomonsson E., Johansson A., Byström M., et al. (2006). Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 59, 1818–1830. 10.1111/j.1365-2958.2006.05061.x [DOI] [PubMed] [Google Scholar]
- Forslund A. L., Salomonsson E. N., Golovliov I., Kuoppa K., Michell S., Titball R., et al. (2010). The type IV pilin, PilA, is required for full virulence of Francisella tularensis subspecies tularensis. BMC Microbiol. 10:227. 10.1186/1471-2180-10-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman Z. N., Dorus S., Waterfield N. R. (2013). The KdpD/KdpE two-component system: integrating K(+) homeostasis and virulence. PLoS Pathog. 9:e1003201. 10.1371/journal.ppat.1003201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller J. R., Craven R. R., Hall J. D., Kijek T. M., Taft-Benz S., Kawula T. H. (2008). RipA, a cytoplasmic membrane protein conserved among Francisella species, is required for intracellular survival. Infect. Immun. 76, 4934–4943. 10.1128/IAI.00475-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller J. R., Kijek T. M., Taft-Benz S., Kawula T. H. (2009). Environmental and intracellular regulation of Francisella tularensis ripA. BMC Microbiol. 9:216. 10.1186/1471-2180-9-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop M., Mastroeni P., Green M., Titball R. W. (2001). Role of antibody to lipopolysaccharide in protection against low- and high-virulence strains of Francisella tularensis. Vaccine 19, 4465–4472. 10.1016/S0264-410X(01)00189-X [DOI] [PubMed] [Google Scholar]
- Fulop M., Webber T., Manchee R. (1993). Activation of the complement system by Francisella tularensis lipopolysaccharide. New Microbiol. 16, 141–147. [PubMed] [Google Scholar]
- Geier H., Celli J. (2011). Phagocytic receptors dictate phagosomal escape and intracellular proliferation of Francisella tularensis. Infect. Immun. 79, 2204–2214. 10.1128/IAI.01382-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil H., Benach J. L., Thanassi D. G. (2004). Presence of pili on the surface of Francisella tularensis. Infect. Immun. 72, 3042–3047. 10.1128/IAI.72.5.3042-3047.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil H., Platz G. J., Forestal C. A., Monfett M., Bakshi C. S., Sellati T. J., et al. (2006). Deletion of TolC orthologs in Francisella tularensis identifies roles in multidrug resistance and virulence. Proc. Natl. Acad. Sci. U.S.A. 103, 12897–12902. 10.1073/pnas.0602582103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goemans C., Denoncin K., Collet J. F. (2014). Folding mechanisms of periplasmic proteins. Biochim. Biophys. Acta 1843, 1517–1528. 10.1016/j.bbamcr.2013.10.014 [DOI] [PubMed] [Google Scholar]
- Gunn J. S., Ernst R. K. (2007). The structure and function of Francisella lipopolysaccharide. Ann. N. Y. Acad. Sci. 1105, 202–218. 10.1196/annals.1409.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A. J., Bolton D. L., Pelletier M. R., Brittnacher M. J., Gallagher L. A., Kaul R., et al. (2006). Type IV pili-mediated secretion modulates Francisella virulence. Mol. Microbiol. 62, 227–237. 10.1111/j.1365-2958.2006.05365.x [DOI] [PubMed] [Google Scholar]
- Hajjar A. M., Harvey M. D., Shaffer S. A., Goodlett D. R., Sjöstedt A., Edebro H., et al. (2006). Lack of in vitro and in vivo recognition of Francisella tularensis subspecies lipopolysaccharide by Toll-like receptors. Infect. Immun. 74, 6730–6738. 10.1128/IAI.00934-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare R. F., Hueffer K. (2014). Francisella novicida pathogenicity island encoded proteins were secreted during infection of macrophage-like cells. PLoS ONE 9:e105773. 10.1371/journal.pone.0105773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley G., Taylor R., Prior J., Newstead S., Hitchen P. G., Morris H. R., et al. (2006). Grey variants of the live vaccine strain of Francisella tularensis lack lipopolysaccharide O-antigen, show reduced ability to survive in macrophages and do not induce protective immunity in mice. Vaccine 24, 989–996. 10.1016/j.vaccine.2005.08.075 [DOI] [PubMed] [Google Scholar]
- Hestvik G., Warns-Petit E., Smith L. A., Fox N. J., Uhlhorn H., Artois M., et al. (2015). The status of tularemia in Europe in a one-health context: a review. Epidemiol. Infect. 143, 2137–2160. 10.1017/S0950268814002398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B. T., Dong T. G., Mekalanos J. J. (2014). A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15, 9–21. 10.1016/j.chom.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood A. M. (1977). Virulence factors of Francisella tularensis. J. Hyg. (Lond). 79, 47–60. 10.1017/S0022172400052840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. T., Mortensen B. L., Taxman D. J., Craven R. R., Taft-Benz S., Kijek T. M., et al. (2010). Deletion of ripA alleviates suppression of the inflammasome and MAPK by Francisella tularensis. J. Immunol. 185, 5476–5485. 10.4049/jimmunol.1002154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley J. F., Conley P. G., Hagman K. E., Norgard M. V. (2007). Characterization of Francisella tularensis outer membrane proteins. J. Bacteriol. 189, 561–574. 10.1128/JB.01505-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley J. F., Conley P. G., Rasko D. A., Hagman K. E., Apicella M. A., Norgard M. V. (2008). Native outer membrane proteins protect mice against pulmonary challenge with virulent type A Francisella tularensis. Infect. Immun. 76, 3664–3671. 10.1128/IAI.00374-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley J. F., Robertson G. T., Norgard M. V. (2010). Method for the isolation of Francisella tularensis outer membranes. J. Vis. Exp. 40:e2044. 10.3791/2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janovská S., Pávková I., Hubálek M., Lenco J., Macela A., Stulík J. (2007). Identification of immunoreactive antigens in membrane proteins enriched fraction from Francisella tularensis LVS. Immunol. Lett. 108, 151–159. 10.1016/j.imlet.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Jia Q., Lee B. Y., Bowen R., Dillon B. J., Som S. M., Horwitz M. A. (2010). A Francisella tularensis live vaccine strain (LVS) mutant with a deletion in capB, encoding a putative capsular biosynthesis protein, is significantly more attenuated than LVS yet induces potent protective immunity in mice against F. tularensis challenge. Infect. Immun. 78, 4341–4355. 10.1128/IAI.00192-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. D., Faron M., Rasmussen J. A., Fletcher J. R. (2014). Uncovering the components of the Francisella tularensis virulence stealth strategy. Front. Cell. Infect. Microbiol. 4:32. 10.3389/fcimb.2014.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadzhaev K., Zingmark C., Golovliov I., Bolanowski M., Shen H., Conlan W., et al. (2009). Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS ONE 4:e5463. 10.1371/journal.pone.0005463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanistanon D., Powell D. A., Hajjar A. M., Pelletier M. R., Cohen I. E., Way S. S., et al. (2012). Role of Francisella lipid A phosphate modification in virulence and long-term protective immune responses. Infect. Immun. 80, 943–951. 10.1128/IAI.06109-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P., Johansson A., Wagner D. M. (2007). Molecular epidemiology, evolution, and ecology of Francisella. Ann. N. Y. Acad. Sci. 1105, 30–66. 10.1196/annals.1409.011 [DOI] [PubMed] [Google Scholar]
- Kieffer T. L., Cowley S., Nano F. E., Elkins K. L. (2003). Francisella novicida LPS has greater immunobiological activity in mice than F. tularensis LPS, and contributes to F. novicida murine pathogenesis. Microbes Infect. 5, 397–403. 10.1016/S1286-4579(03)00052-2 [DOI] [PubMed] [Google Scholar]
- Kingry L. C., Petersen J. M. (2014). Comparative review of Francisella tularensis and Francisella novicida. Front. Cell. Infect. Microbiol. 4:35. 10.3389/fcimb.2014.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konecna K., Hernychova L., Reichelova M., Lenco J., Klimentova J., Stulik J., et al. (2010). Comparative proteomic profiling of culture filtrate proteins of less and highly virulent Francisella tularensis strains. Proteomics 10, 4501–4511. 10.1002/pmic.201000248 [DOI] [PubMed] [Google Scholar]
- Lai X. H., Shirley R. L., Crosa L., Kanistanon D., Tempel R., Ernst R. K., et al. (2010). Mutations of Francisella novicida that alter the mechanism of its phagocytosis by murine macrophages. PLoS ONE 5:e11857. 10.1371/journal.pone.0011857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson P., Oyston P. C., Chain P., Chu M. C., Duffield M., Fuxelius H. H., et al. (2005). The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37, 153–159. 10.1038/ng1499 [DOI] [PubMed] [Google Scholar]
- Lee B. Y., Horwitz M. A., Clemens D. L. (2006). Identification, recombinant expression, immunolocalization in macrophages, and T-cell responsiveness of the major extracellular proteins of Francisella tularensis. Infect. Immun. 74, 4002–4013. 10.1128/IAI.00257-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Powell D. A., Shaffer S. A., Rasko D. A., Pelletier M. R., Leszyk J. D., et al. (2012). LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, 8716–8721. 10.1073/pnas.1202908109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann S. R., Peng K., Long M. E., Hunt J. R., Apicella M. A., Monack D. M., et al. (2011). Francisella tularensis Schu S4 O-antigen and capsule biosynthesis gene mutants induce early cell death in human macrophages. Infect. Immun. 79, 581–594. 10.1128/IAI.00863-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Honn M., Golovlev I., Kadzhaev K., Conlan W., Sjöstedt A. (2009). The 58-kilodalton major virulence factor of Francisella tularensis is required for efficient utilization of iron. Infect. Immun. 77, 4429–4436. 10.1128/IAI.00702-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Shen H., Zingmark C., Golovliov I., Conlan W., Sjöstedt A. (2007). Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect. Immun. 75, 1303–1309. 10.1128/IAI.01717-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludu J. S., de Bruin O. M., Duplantis B. N., Schmerk C. L., Chou A. Y., Elkins K. L., et al. (2008). The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J. Bacteriol. 190, 4584–4595. 10.1128/JB.00198-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J., Yu Z., Wagner S., de Gier J. W. (2012). Biogenesis of inner membrane proteins in Escherichia coli. Biochim. Biophys. Acta 1817, 965–976. 10.1016/j.bbabio.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Mahawar M., Atianand M. K., Dotson R. J., Mora V., Rabadi S. M., Metzger D. W., et al. (2012). Identification of a novel Francisella tularensis factor required for intramacrophage survival and subversion of innate immune response. J. Biol. Chem. 287, 25216–25229. 10.1074/jbc.M112.367672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahawar M., Rabadi S. M., Banik S., Catlett S. V., Metzger D. W., Malik M., et al. (2013). Identification of a live attenuated vaccine candidate for tularemia prophylaxis. PLoS ONE 8:e61539. 10.1371/journal.pone.0061539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Garcia J. M., Hansen D. T., Zook J., Loskutov A. V., Robida M. D., Craciunescu F. M., et al. (2014). Purification and biophysical characterization of the CapA membrane protein FTT0807 from Francisella tularensis. Biochemistry 53, 1958–1970. 10.1021/bi401644s [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig W. D., Koller A., Thanassi D. G. (2013). Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J. Bacteriol. 195, 1120–1132. 10.1128/JB.02007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre P. B., O'Brien K. L., Greenwood B., van de Beek D. (2012). Effect of vaccines on bacterial meningitis worldwide. Lancet 380, 1703–1711. 10.1016/S0140-6736(12)61187-8 [DOI] [PubMed] [Google Scholar]
- McRae S., Pagliai F. A., Mohapatra N. P., Gener A., Mahmou A. S., Gunn J. S., et al. (2010). Inhibition of AcpA phosphatase activity with ascorbate attenuates Francisella tularensis intramacrophage survival. J. Biol. Chem. 285, 5171–5177. 10.1074/jbc.M109.039511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo A. A., Mahawar M., Sellati T. J., Malik M., Metzger D. W., Melendez J. A., et al. (2009). Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J. Bacteriol. 191, 6447–6456. 10.1128/JB.00534-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo A., Sledjeski D. D., Lipski S., Wooten R. M., Basrur V., Lafontaine E. R. (2006). Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS Microbiol. Lett. 263, 102–108. 10.1111/j.1574-6968.2006.00413.x [DOI] [PubMed] [Google Scholar]
- Merdanovic M., Clausen T., Kaiser M., Huber R., Ehrmann M. (2011). Protein quality control in the bacterial periplasm. Annu. Rev. Microbiol. 65, 149–168. 10.1146/annurev-micro-090110-102925 [DOI] [PubMed] [Google Scholar]
- Michell S. L., Dean R. E., Eyles J. E., Hartley M. G., Waters E., Prior J. L., et al. (2010). Deletion of the Bacillus anthracis capB homologue in Francisella tularensis subspecies tularensis generates an attenuated strain that protects mice against virulent tularaemia. J. Med. Microbiol. 59, 1275–1284. 10.1099/jmm.0.018911-0 [DOI] [PubMed] [Google Scholar]
- Miller C. N., Steele S. P., Brunton J. C., Jenkins R. J., LoVullo E. D., Taft-Benz S. A., et al. (2014). Extragenic suppressor mutations in ΔripA disrupt stability and function of LpxA. BMC Microbiol. 14:336. 10.1186/s12866-014-0336-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. I., Ernst R. K., Bader M. W. (2005). LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 3, 36–46. 10.1038/nrmicro1068 [DOI] [PubMed] [Google Scholar]
- Mohapatra N. P., Balagopal A., Soni S., Schlesinger L. S., Gunn J. S. (2007). AcpA is a Francisella acid phosphatase that affects intramacrophage survival and virulence. Infect. Immun. 75, 390–396. 10.1128/IAI.01226-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra N. P., Soni S., Rajaram M. V., Dang P. M., Reilly T. J., El-Benna J., et al. (2010). Francisella acid phosphatases inactivate the NADPH oxidase in human phagocytes. J. Immunol. 184, 5141–5150. 10.4049/jimmunol.0903413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra N. P., Soni S., Reilly T. J., Liu J., Klose K. E., Gunn J. S. (2008). Combined deletion of four Francisella novicida acid phosphatases attenuates virulence and macrophage vacuolar escape. Infect. Immun. 76, 3690–3699. 10.1128/IAI.00262-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrievich A. N., Kondakova A. N., Valade E., Platonov M. E., Vakhrameeva G. M., Shaikhutdinova R. Z., et al. (2010). Biological properties and structure of the lipopolysaccharide of a vaccine strain of Francisella tularensis generated by inactivation of a quorum sensing system gene qseC. Biochemistry Mosc. 75, 443–451. 10.1134/S0006297910040073 [DOI] [PubMed] [Google Scholar]
- Molins C. R., Delorey M. J., Yockey B. M., Young J. W., Belisle J. T., Schriefer M. E., et al. (2014). Virulence difference between the prototypic Schu S4 strain (A1a) and Francisella tularensis A1a, A1b, A2 and type B strains in a murine model of infection. BMC Infect. Dis. 14:67. 10.1186/1471-2334-14-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molins C. R., Delorey M. J., Yockey B. M., Young J. W., Sheldon S. W., Reese S. M., et al. (2010). Virulence differences among Francisella tularensis subsp. tularensis clades in mice. PLoS ONE 5:e10205. 10.1371/journal.pone.0010205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen B. L., Fuller J. R., Taft-Benz S., Collins E. J., Kawula T. H. (2012). Francisella tularensis RipA protein topology and identification of functional domains. J. Bacteriol. 194, 1474–1484. 10.1128/JB.06327-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moule M. G., Monack D. M., Schneider D. S. (2010). Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 6:e1001065. 10.1371/journal.ppat.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallaparaju K. C., Yu J. J., Rodriguez S. A., Zogaj X., Manam S., Guentzel M. N., et al. (2011). Evasion of IFN-gamma signaling by Francisella novicida is dependent upon Francisella outer membrane protein C. PLoS ONE 6:e18201. 10.1371/journal.pone.0018201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J. Q., Gilley R. P., Zogaj X., Rodriguez S. A., Klose K. E. (2014). Lipidation of the FPI protein IglE contributes to Francisella tularensis ssp. novicida intramacrophage replication and virulence. Pathog. Dis. 72, 10–18. 10.1111/2049-632X.12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okan N. A., Kasper D. L. (2013). The atypical lipopolysaccharide of Francisella. Carbohydr. Res. 378, 79–83. 10.1016/j.carres.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyston P. C. (2009). Francisella tularensis vaccines. Vaccine 27(Suppl 4), D48–D51. 10.1016/j.vaccine.2009.07.090 [DOI] [PubMed] [Google Scholar]
- Oyston P. C., Sjostedt A., Titball R. W. (2004). Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2, 967–978. 10.1038/nrmicro1045 [DOI] [PubMed] [Google Scholar]
- Parra M. C., Shaffer S. A., Hajjar A. M., Gallis B. M., Hager A., Goodlett D. R., et al. (2010). Identification, cloning, expression, and purification of Francisella lpp3: an immunogenic lipoprotein. Microbiol. Res. 165, 531–545. 10.1016/j.micres.2009.11.004 [DOI] [PubMed] [Google Scholar]
- Pavkova I., Hubálek M., Zechovská J., Lenco J., Stulík J. (2005). Francisella tularensis live vaccine strain: proteomic analysis of membrane proteins enriched fraction. Proteomics 5, 2460–2467. 10.1002/pmic.200401213 [DOI] [PubMed] [Google Scholar]
- Pavkova I., Reichelova M., Larsson P., Hubalek M., Vackova J., Forsberg A., et al. (2006). Comparative proteome analysis of fractions enriched for membrane-associated proteins from Francisella tularensis subsp. tularensis and F. tularensis subsp. holarctica strains. J. Proteome Res. 5, 3125–3134. 10.1021/pr0601887 [DOI] [PubMed] [Google Scholar]
- Peng K., Broz P., Jones J., Joubert L. M., Monack D. (2011). Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell. Microbiol. 13, 1586–1600. 10.1111/j.1462-5822.2011.01643.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J. M., Molins C. R. (2010). Subpopulations of Francisella tularensis ssp. tularensis and holarctica: identification and associated epidemiology. Future Microbiol. 5, 649–661. 10.2217/fmb.10.17 [DOI] [PubMed] [Google Scholar]
- Phillips N. J., Schilling B., McLendon M. K., Apicella M. A., Gibson B. W. (2004). Novel modification of lipid A of Francisella tularensis. Infect. Immun. 72, 5340–5348. 10.1128/IAI.72.9.5340-5348.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. (2006). Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 4, 629–636. 10.1038/nrmicro1464 [DOI] [PubMed] [Google Scholar]
- Pierson T., Matrakas D., Taylor Y. U., Manyam G., Morozov V. N., Zhou W., et al. (2011). Proteomic characterization and functional analysis of outer membrane vesicles of Francisella novicida suggests possible role in virulence and use as a vaccine. J. Proteome Res. 10, 954–967. 10.1021/pr1009756 [DOI] [PubMed] [Google Scholar]
- Platz G. J., Bublitz D. C., Mena P., Benach J. L., Furie M. B., Thanassi D. G. (2010). A tolC mutant of Francisella tularensis is hypercytotoxic compared to the wild type and elicits increased proinflammatory responses from host cells. Infect. Immun. 78, 1022–1031. 10.1128/IAI.00992-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pore D., Chakrabarti M. K. (2013). Outer membrane protein A (OmpA) from Shigella flexneri 2a: a promising subunit vaccine candidate. Vaccine 31, 3644–3650. 10.1016/j.vaccine.2013.05.100 [DOI] [PubMed] [Google Scholar]
- Preston J. A., Dockrell D. H. (2008). Virulence factors in pneumococcal respiratory pathogenesis. Future Microbiol. 3, 205–221. 10.2217/17460913.3.2.205 [DOI] [PubMed] [Google Scholar]
- Prior J. L., Prior R. G., Hitchen P. G., Diaper H., Griffin K. F., Morris H. R., et al. (2003). Characterization of the O antigen gene cluster and structural analysis of the O antigen of Francisella tularensis subsp. tularensis. J. Med. Microbiol. 52, 845–851. 10.1099/jmm.0.05184-0 [DOI] [PubMed] [Google Scholar]
- Qin A., Mann B. J. (2006). Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 6:69. 10.1186/1471-2180-6-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A., Scott D. W., Rabideau M. M., Moore E. A., Mann B. J. (2011). Requirement of the CXXC motif of novel Francisella infectivity potentiator protein B FipB, and FipA in virulence of F. tularensis subsp. tularensis. PLoS ONE 6:e24611. 10.1371/journal.pone.0024611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A., Scott D. W., Thompson J. A., Mann B. J. (2009). Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect Immun 77, 152–161. 10.1128/IAI.01113-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin A., Zhang Y., Clark M. E., Rabideau M. M., Millan Barea L. R., Mann B. J. (2014). FipB, an essential virulence factor of Francisella tularensis subsp. tularensis, has dual roles in disulfide bond formation. J. Bacteriol. 196, 3571–3581. 10.1128/JB.01359-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G., Meeker A., Dragulev B. (2008). fslE is necessary for siderophore-mediated iron acquisition in Francisella tularensis Schu S4. J. Bacteriol. 190, 5353–5361. 10.1128/JB.00181-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G., Sen B. (2014). The FupA/B protein uniquely facilitates transport of ferrous iron and siderophore-associated ferric iron across the outer membrane of Francisella tularensis live vaccine strain. Microbiology (Reading Engl). 160, 446–457. 10.1099/mic.0.072835-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan G., Sen B., Johnson R. (2012). Paralogous outer membrane proteins mediate uptake of different forms of iron and synergistically govern virulence in Francisella tularensis tularensis. J. Biol. Chem. 287, 25191–25202. 10.1074/jbc.M112.371856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Moreira C. G., Li de R., Reading N. C., Ritchie J. M., Waldor M. K., et al. (2008). Targeting QseC signaling and virulence for antibiotic development. Science 321, 1078–1080. 10.1126/science.1160354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen J. A., Post D. M., Gibson B. W., Lindemann S. R., Apicella M. A., Meyerholz D. K., et al. (2014). Francisella tularensis Schu S4 lipopolysaccharide core sugar and O-antigen mutants are attenuated in a mouse model of tularemia. Infect. Immun. 82, 1523–1539. 10.1128/IAI.01640-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond B., Young J. C., Pallett M., Endres R. G., Clements A., Frankel G. (2013). Subversion of trafficking, apoptosis, and innate immunity by type III secretion system effectors. Trends Microbiol. 21, 430–441. 10.1016/j.tim.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Rebeil R., Ernst R. K., Jarrett C. O., Adams K. N., Miller S. I., Hinnebusch B. J. (2006). Characterization of late acyltransferase genes of Yersinia pestis and their role in temperature-dependent lipid A variation. J. Bacteriol. 188, 1381–1388. 10.1128/JB.188.4.1381-1388.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly T. J., Baron G. S., Nano F. E., Kuhlenschmidt M. S. (1996). Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J. Biol. Chem. 271, 10973–10983. 10.1074/jbc.271.18.10973 [DOI] [PubMed] [Google Scholar]
- Ren G., Champion M. M., Huntley J. F. (2014). Identification of disulfide bond isomerase substrates reveals bacterial virulence factors. Mol. Microbiol. 94, 926–944. 10.1111/mmi.12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G. T., Case E. D., Dobbs N., Ingle C., Balaban M., Celli J., et al. (2014). FTT0831c/FTL_0325 contributes to Francisella tularensis cell division, maintenance of cell shape, and structural integrity. Infect. Immun. 82, 2935–2948. 10.1128/IAI.00102-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G. T., Child R., Ingle C., Celli J., Norgard M. V. (2013). IglE is an outer membrane-associated lipoprotein essential for intracellular survival and murine virulence of type A Francisella tularensis. Infect. Immun. 81, 4026–4040. 10.1128/IAI.00595-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B., Peterson S. B., Mougous J. D. (2014). Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148. 10.1038/nrmicro3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonsson E., Kuoppa K., Forslund A. L., Zingmark C., Golovliov I., Sjöstedt A., et al. (2009). Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infect. Immun. 77, 3424–3431. 10.1128/IAI.00196-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström G., Löfgren S., Tärnvik A. (1988). A capsule-deficient mutant of Francisella tularensis LVS exhibits enhanced sensitivity to killing by serum but diminished sensitivity to killing by polymorphonuclear leukocytes. Infect. Immun. 56, 1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström G., Sjöstedt A., Johansson T., Kuoppa K., Williams J. C. (1992). Immunogenicity and toxicity of lipopolysaccharide from Francisella tularensis LVS. FEMS Microbiol. Immunol. 5, 201–210. 10.1111/j.1574-6968.1992.tb05902.x [DOI] [PubMed] [Google Scholar]
- Schilling B., McLendon M. K., Phillips N. J., Apicella M. A., Gibson B. W. (2007). Characterization of lipid A acylation patterns in Francisella tularensis, Francisella novicida, and Francisella philomiragia using multiple-stage mass spectrometry and matrix-assisted laser desorption/ionization on an intermediate vacuum source linear ion trap. Anal. Chem. 79, 1034–1042. 10.1021/ac061654e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Klimentova J., Rehulka P., Straskova A., Spidlova P., Szotakova B., et al. (2013). Francisella tularensis subsp. holarctica DsbA homologue: a thioredoxin-like protein with chaperone function. Microbiology 159, 2364–2374. 10.1099/mic.0.070516-0 [DOI] [PubMed] [Google Scholar]
- Sen B., Meeker A., Ramakrishnan G. (2010). The fslE homolog, FTL_0439 (fupA/B), mediates siderophore-dependent iron uptake in Francisella tularensis LVS. Infect. Immun. 78, 4276–4285. 10.1128/IAI.00503-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senitkova I., Spidlova P., Stulik J. (2015). Cooperation of both, the FKBP_N-like and the DSBA-like, domains is necessary for the correct function of FTS_1067 protein involved in Francisella tularensis virulence and pathogenesis. Pathog. Dis. 73:ftv030. 10.1093/femspd/ftv030 [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Kahne D., Walker S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414. 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöstedt A. (2006). Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 8, 561–567. 10.1016/j.micinf.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Soni S., Ernst R. K., Muszynski A., Mohapatra N. P., Perry M. B., Vinogradov E., et al. (2010). Francisella tularensis blue-gray phase variation involves structural modifications of lipopolysaccharide o-antigen, core and lipid a and affects intramacrophage survival and vaccine efficacy. Front. Microbiol. 1:129. 10.3389/fmicb.2010.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin V. M., Pavlovich N. V., Prozorova L. A. (1996). Francisella tularensis resistance to bactericidal action of normal human serum. FEMS Immunol. Med. Microbiol. 13, 249–252. 10.1111/j.1574-695X.1996.tb00246.x [DOI] [PubMed] [Google Scholar]
- Straskova A., Pavkova I., Link M., Forslund A. L., Kuoppa K., Noppa L., et al. (2009). Proteome analysis of an attenuated Francisella tularensis dsbA mutant: identification of potential DsbA substrate proteins. J. Proteome Res. 8, 5336–5346. 10.1021/pr900570b [DOI] [PubMed] [Google Scholar]
- Su J., Yang J., Zhao D., Kawula T. H., Banas J. A., Zhang J. R. (2007). Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75, 3089–3101. 10.1128/IAI.01865-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Kagan J. C. (2014). A cross-disciplinary perspective on the innate immune responses to bacterial lipopolysaccharide. Mol. Cell 54, 212–223. 10.1016/j.molcel.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakran S., Li H., Lavine C. L., Miller M. A., Bina J. E., Bina X. R., et al. (2008). Identification of Francisella tularensis lipoproteins that stimulate the toll-like receptor (TLR) 2/TLR1 heterodimer. J. Biol. Chem. 283, 3751–3760. 10.1074/jbc.M706854200 [DOI] [PubMed] [Google Scholar]
- Thomas R. M., Titball R. W., Oyston P. C., Griffin K., Waters E., Hitchen P. G., et al. (2007). The immunologically distinct O antigens from Francisella tularensis subspecies tularensis and Francisella novicida are both virulence determinants and protective antigens. Infect. Immun. 75, 371–378. 10.1128/IAI.01241-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Holland I. B., Schmitt L. (2014). The Type 1 secretion pathway - the hemolysin system and beyond. Biochim. Biophys. Acta 1843, 1629–1641. 10.1016/j.bbamcr.2013.09.017 [DOI] [PubMed] [Google Scholar]
- Twine S., Byström M., Chen W., Forsman M., Golovliov I., Johansson A., et al. (2005a). A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58-kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect. Immun. 73, 8345–8352. 10.1128/IAI.73.12.8345-8352.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twine S. M., Mykytczuk N. C., Petit M., Tremblay T. L., Lanthier P., Conlan J. W., et al. (2005b). Francisella tularensis proteome: low levels of ASB-14 facilitate the visualization of membrane proteins in total protein extracts. J. Proteome Res. 4, 1848–1854. 10.1021/pr050102u [DOI] [PubMed] [Google Scholar]
- Twine S. M., Vinogradov E., Lindgren H., Sjostedt A., Conlan J. W. (2012). Roles for wbtC, wbtI, and kdtA genes in lipopolysaccharide biosynthesis, protein glycosylation, virulence, and immunogenicity in strain SCHU S4. Pathogens 1, 12–29. 10.3390/pathogens1010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov E., Perry M. B. (2004). Characterisation of the core part of the lipopolysaccharide O-antigen of Francisella novicida (U112). Carbohydr. Res. 339, 1643–1648. 10.1016/j.carres.2004.04.013 [DOI] [PubMed] [Google Scholar]
- Vinogradov E., Perry M. B., Conlan J. W. (2002). Structural analysis of Francisella tularensis lipopolysaccharide. Eur. J. Biochem. 269, 6112–6118. 10.1046/j.1432-1033.2002.03321.x [DOI] [PubMed] [Google Scholar]
- Wang Q., Shi X., Leymarie N., Madico G., Sharon J., Costello C. E., et al. (2011). A typical preparation of Francisella tularensis O-antigen yields a mixture of three types of saccharides. Biochemistry 50, 10941–10950. 10.1021/bi201450v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Karbarz M. J., McGrath S. C., Cotter R. J., Raetz C. R. (2004). MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of Francisella novicida LpxE expressed in Escherichia coli. J. Biol. Chem. 279, 49470–49478. 10.1074/jbc.M409078200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ribeiro A. A., Guan Z., Abraham S. N., Raetz C. R. (2007). Attenuated virulence of a Francisella mutant lacking the lipid A 4′-phosphatase. Proc. Natl. Acad. Sci. U.S.A. 104, 4136–4141. 10.1073/pnas.0611606104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ribeiro A. A., Guan Z., McGrath S. C., Cotter R. J., Raetz C. R. (2006). Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry 45, 14427–14440. 10.1021/bi061767s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrly T. D., Chong A., Virtaneva K., Sturdevant D. E., Child R., Edwards J. A., et al. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150. 10.1111/j.1462-5822.2009.01316.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel W. A., Demuth D. R. (2015). QseBC, a two-component bacterial adrenergic receptor and global regulator of virulence in Enterobacteriaceae and Pasteurellaceae. Mol. Oral Microbiol. [Epub ahead of print]. 10.1111/omi.12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D. S., Brotcke A., Henry T., Margolis J. J., Chan K., Monack D. M. (2007). In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U.S.A. 104, 6037–6042. 10.1073/pnas.0609675104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. (2006). Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68. 10.1146/annurev.biochem.75.103004.142545 [DOI] [PubMed] [Google Scholar]
- Willis L. M., Whitfield C. (2013). Structure, biosynthesis, and function of bacterial capsular polysaccharides synthesized by ABC transporter-dependent pathways. Carbohydr. Res. 378, 35–44. 10.1016/j.carres.2013.05.007 [DOI] [PubMed] [Google Scholar]
- Zhao J., Raetz C. R. (2010). A two-component Kdo hydrolase in the inner membrane of Francisella novicida. Mol. Microbiol. 78, 820–836. 10.1111/j.1365-2958.2010.07305.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X., Chakraborty S., Liu J., Thanassi D. G., Klose K. E. (2008). Characterization of the Francisella tularensis subsp. novicida type IV pilus. Microbiology 154, 2139–2150. 10.1099/mic.0.2008/018077-0 [DOI] [PubMed] [Google Scholar]
- Zook J., Mo G., Sisco N. J., Craciunescu F. M., Hansen D. T., Baravati B., et al. (2015). NMR structure of Francisella tularensis virulence determinant reveals structural homology to Bet v1 allergen proteins. Structure 23, 1116–1122. 10.1016/j.str.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]