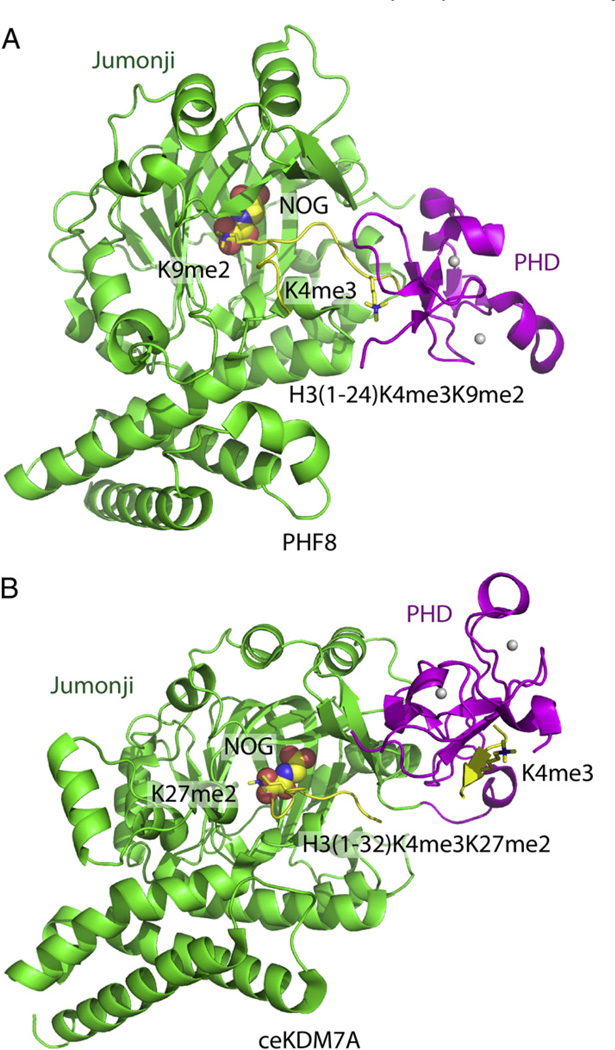
Fig. 4.
Structural basis for recognition by histone modification-directed histone modification enzyme. (A) Ribbon representation of the crystal structure of PHF8 in complex with H3(1–24)K4me3/K9me2 peptide (PDB code: 3KV4). The PHD finger, Jumonji domain and the bound peptide are colored in magenta, green and yellow, respectively. The NOG cofactor is shown in a space-filling representation. The H3K4m3mark, which is specifically recognized by the PHD finger, and the H3K9me2 mark, which specifically inserts into the active site of the Jumonji domain, are highlighted in stick representations. The PHF8 enzyme shows a bent conformation upon K4me3 binding into the PHD pocket, with the distance between the PHD pocket and Jumonji active site short enough to allow the H3K9me2 mark to simultaneously insert into the Jumonji active site. (B) Ribbon representation of the crystal structure of ceKDM7A in complex with H3(1–32)K4me3/K27me2 peptide (PDB code: 3N9P). The PHD finger, Jumonji domain and the bound peptide are colored in magenta, green and yellow, respectively. The NOG cofactor is shown in a space filling representation. The H3K4me3 mark, which is specifically recognized by the PHD finger, and the H3K27me2 mark, which specifically inserts into the active site of the Jumonji domain, are highlighted in stick representation. The ceKDM7A enzyme shows an extended conformation upon K4me3 binding into the PHD pocket, with the distance between the PHD pocket and Jumonji active site being too long to allow simultaneous recognition of H3K9me2, but should allow simultaneous recognition of H3K27me2 mark by inserting it into the Jumonji active site.