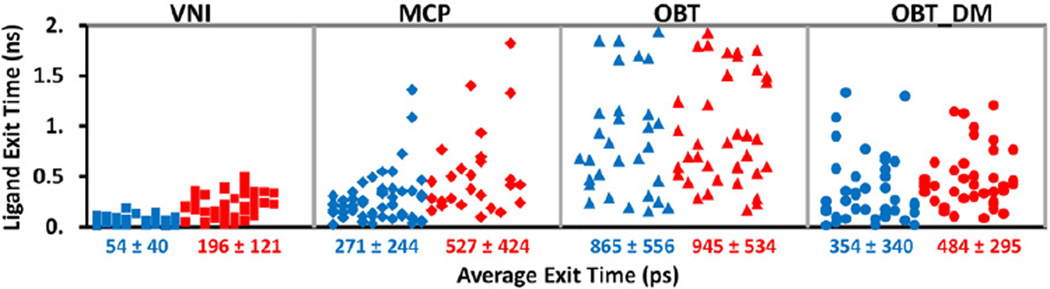
Fig. 5.
The times required for the four ligands to egress through tunnel 2 f in T. brucei CYP51 (blue: soluble model, red: membrane-bound model) in each RAMD trajectory. Each symbol represents the result of one RAMD trajectory and the average exit times of all simulations with exit through tunnel 2 f are given below the x-axis in ps. Times obtained in RAMD simulations are expected to be much shorter than the real exit times but their relative values indicate the extent to which ligand egress from the active site is hindered; longer average times correspond to higher barriers to ligand egress.