Abstract
Riboswitches are metabolite-sensing RNAs, typically located in the non-coding portions of messenger RNAs, that control the synthesis of metabolite-related proteins1–3. Here we describe a 2.05 Å crystal structure of a riboswitch domain from the Escherichia coli thiM mRNA4 that responds to the coenzyme thiamine pyrophosphate (TPP). TPP is an active form of vitamin B1, an essential participant in many protein-catalysed reactions5. Organisms from all three domains of life6–9, including bacteria, plants and fungi, use TPP-sensing riboswitches to control genes responsible for importing or synthesizing thiamine and its phosphorylated derivatives, making this riboswitch class the most widely distributed member of the metabolite-sensing RNA regulatory system. The structure reveals a complex folded RNA in which one subdomain forms an intercalation pocket for the 4-amino-5-hydroxymethyl-2-methyl-pyrimidine moiety of TPP, whereas another subdomain forms a wider pocket that uses bivalent metal ions and water molecules to make bridging contacts to the pyrophosphate moiety of the ligand. The two pockets are positioned to function as a molecular measuring device that recognizes TPP in an extended conformation. The central thiazole moiety is not recognized by the RNA, which explains why the antimicrobial compound pyrithiamine pyrophosphate targets this riboswitch and downregulates the expression of thiamine metabolic genes. Both the natural ligand and its drug-like analogue stabilize secondary and tertiary structure elements that are harnessed by the riboswitch to modulate the synthesis of the proteins coded by the mRNA. In addition, this structure provides insight into how folded RNAs can form precision binding pockets that rival those formed by protein genetic factors.
More than 2% of the genes in some species are regulated by riboswitches4,10–13, whose representative classes compete in number with known metabolite-sensing regulatory proteins14. On metabolite docking, the sensing domain of the riboswitch shows conformational stabilization, which typically alters the base-pairing arrangements in the adjoining expression platform carrying gene-expression signals1–3. Riboswitches have a remarkable affinity for their cognate ligands and can discriminate against even closely related analogues, as shown by biochemical experiments1,13,15 and X-ray structures of purine-sensing riboswitches bound to hypoxanthine16, guanine17 and adenine17. TPP and pyrithiamine pyrophosphate (PTPP; Fig. 1a) are chemically more diverse than purines and pose several additional challenges for negatively charged RNA receptors, namely their large size, conformational flexibility and, most importantly, the presence of negatively charged phosphate groups18.
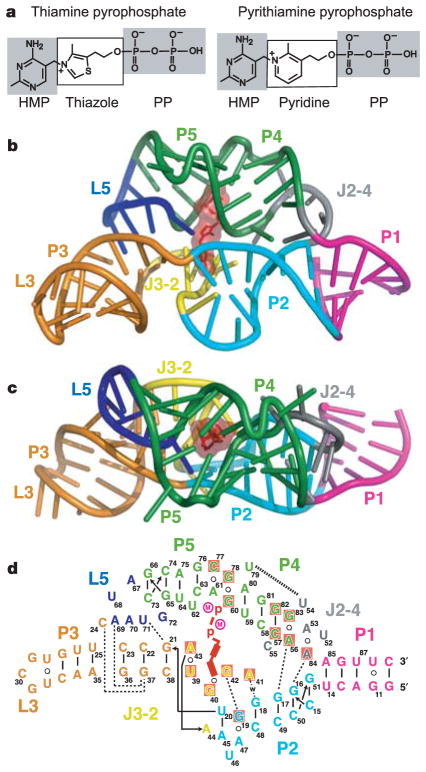
Figure 1. Structural models of a TPP riboswitch and its ligands.
a, Chemical structures of the natural metabolite TPP and the antimicrobial compound PTPP. b, c, Crystal structure of the TPP-bound sensing domain, showing front (b) and top (c) views. The RNA is in a stick-and-ribbon representation, with bound TPP in red. Stems, loops and junctions are colour coded. d, Schematic depiction of the RNA tertiary fold observed in the structure. Tertiary contacts formed by hydrogen bonds (w, water-mediated bonds) between bases and stacking interactions are represented by thin and thick dashed lines, respectively. Red shading shows nucleotides conserved in more than 97% of sequences (J. E. Barrick and R.R.B., unpublished observations). Encircled M notations represent Mg2+ ions.
We crystallized an 80-nucleotide TPP-sensing domain from the thiM mRNA4. This RNA conforms well with the consensus sequence and secondary structure model for TPP riboswitches that was established by previous phylogenetic6–9 and biochemical4,6,19 analyses (Supplementary Fig. S1). The structure (Fig. 1b–d, Supplementary Fig. S2 and Supplementary Table S1) reveals that the conserved nucleotides and secondary structure elements common to all TPP riboswitches form a complex tertiary architecture consisting of two parallel helical domains (P2/J3-2/P3/L3 and J2-4/P4/P5/L5) connected to a helix (P1) by means of a three-way junction.
Unlike most other ligand–RNA interactions that exploit the helical grooves of RNA, TPP is positioned perpendicular to the two main helical domains and uses separate pockets formed by each helical segment to grip the ends of the compound in an extended conformation (Fig. 2). In addition to the interdomain bridge formed by the ligand, the riboswitch uses tertiary contacts between L5 and P3, and between J2-4, P2 and P4, to assist in stabilizing the global fold.
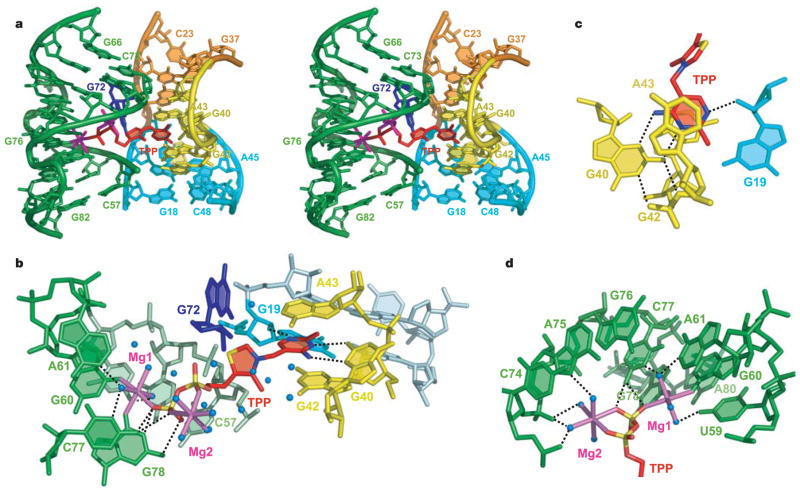
Figure 2. Structure and interactions in the TPP-binding pocket.
a, Stereo view of the central region of the complex containing bound TPP. b, View of TPP, coordinated Mg2+ ions (magenta) and water (blue spheres) in the binding pocket. c, Details of the interactions between the HMP ring and RNA. d, Hydrogen bonding between Mg2+ ions and RNA.
The J3-2 region (Fig. 1d) is essential in the recognition of the 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) ring. The ring intercalates between G42 and A43, and forms hydrogen bonds between its polar functionalities, previously shown to be critical for molecular recognition4, and the G40 base and the 2-OH′ of G19 (Fig. 2b, c). This binding arrangement is held by a T-loop-like turn20 formed by the conserved U39-G40-A41-G42-A43 segment, closed by a reverse Hoogsteen U39·A43 pair (Fig. 3a). The backbone is extended at both G40-A41 and G42-A43, resulting in the mutual intercalation and insertion of the HMP ring between these interacting segments. Further, the fold is stabilized by continuous stacking of A41, G42, HMP, A43 and G21, and by the formation of (G19·A47)·G42 and (G18-C48)·A41 triples (Fig. 2a and Supplementary Fig. S3).
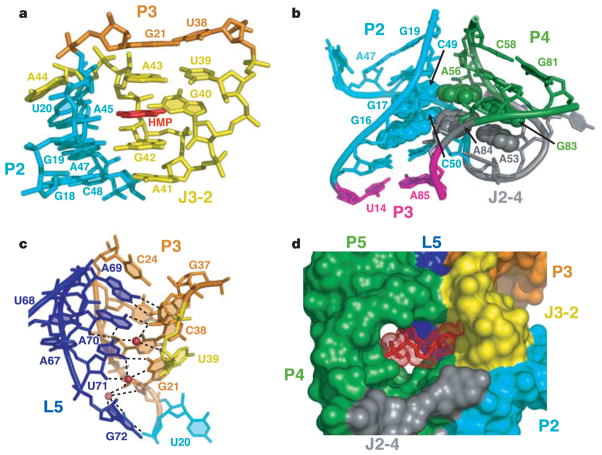
Figure 3. Tertiary interactions defining TPP riboswitch structure and accessibility to the binding pocket.
a, Interaction between J3/2 and P2, mediated by the HMP ring. b, Stabilization of the J2-4 junction by two stacked tetrads (in space-filling representation). c, Interactions between L5 and P3 mediated by three K+ ions (red spheres). d, Surface representation of RNA and accessibility to the TPP-binding pocket. TPP is depicted in a stick and mesh representation.
TPP riboswitches were the first of several classes found to make productive interactions with negatively charged phosphate groups4,12,15. The pyrophosphate group of TPP is bound in a spacious pocket by a pair of hexa-coordinated Mg2+ ions (Mg1 and Mg2) with octahedral ligation geometry that are positioned by direct and water-mediated hydrogen bonds with RNA (Fig. 2d and Supplementary Fig. S1). The terminal phosphate of TPP is coordinated to both Mg1 and Mg2, whereas the thiazole-linked phosphate is coordinated to Mg2. Mg1 is directly coordinated to the O6 carbonyls of the conserved G60 and G78 that are located in the region previously implicated in pyrophosphate recognition4 (Fig. 2b, d, and Supplementary Fig. S2). In addition, the conserved C77 and G78 form hydrogen bonds to the oxygen atoms of the terminal phosphate of TPP.
An analysis of 72 structures of proteins bound to TPP or its analogues from the Protein Data Bank indicates that proteins typically use charged amino acids to position Mg2+, Ca2+ or Mn2+ ions in a site equivalent to Mg2. However, the Mg1 ion is unique to TPP riboswitches. A bivalent cation at this location allows TPP to reach into the pyrophosphate-binding pocket, thereby stabilizing tertiary interactions important for gene regulation and corroborating the requirement of Mg2+ for TPP binding in eukaryotic21 and bacterial (Supplementary Fig. S4) TPP riboswitches. One could consider the ligand as TPP with two bound Mg2+ ions, which overcomes the ligand’s strongly negative electrostatic character. Other riboswitches could use similar simple structural arrangements in combination with cations and water to bind phosphorylated ligands selectively, which would make RNA surprisingly adept at binding negatively charged ligands. The dual bivalent cation arrangement is also distinct from that found in ribosomal RNAs that bind non-phosphorylated antibiotics, in which some contacts with RNA are mediated by single cations22–24.
Nucleotides distal to the ligand-binding sites, conserved in sequence or secondary structure, are important in forming the overall architecture of the TPP-sensing domain. J2-4 (Fig. 1d) is stabilized through the formation of a pair of stacked tetrads, where highly conserved A56·G83 and A53·A84 non-canonical pairs (Supplementary Fig. S5a, b) are aligned in the minor groove of adjacent G·C pairs within P2, forming type I A-minor motifs25. Continuous stacking is observed between P1 and P2, and between non-canonical pairs in J2-4 and P4 (Fig. 3b). This structural arrangement is expected to be stabilized by ligand binding, and reinforces previous suggestions4,12,13 that stabilization of P1 stems in riboswitches is important for the control of gene expression.
Another key tertiary interface involves multiple base–base and base–backbone interactions between nucleotides of L5 and P3 (Fig. 3c), thereby locking into register side-by-side arrangements of P4/P5 and P2/J3-2/P3 segments (Fig. 1b). Three adjacent K+ ions additionally stabilize the interactions (Fig. 3c). It should be noted that nucleotides of the L5/P3 interface are poorly conserved, and other TPP riboswitches could be stabilized by alternative tertiary contacts spanning this region.
A surface view of the complex shows that the thiazole and diphosphate moieties of TPP are visible, whereas the intercalated HMP ring is buried (Fig. 3d). The deep insertion of the HMP ring most probably implies a conformational transition within the riboswitch upon binding TPP. Support for this interpretation comes from in-line probing experiments, which have identified numerous internucleotide linkages that become conformationally restricted on TPP binding4. The structure shows that these regions either contact the ligand directly or participate in critical contacts that form the global structure of the sensing domain.
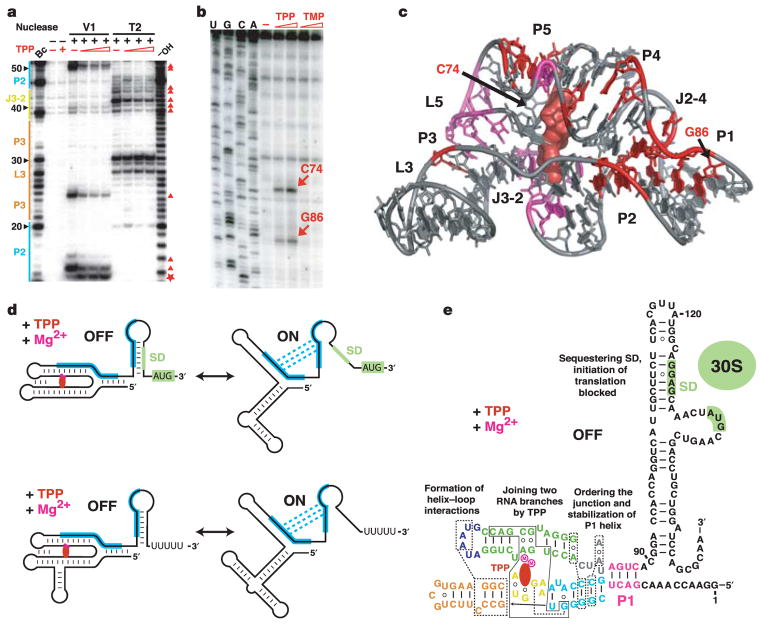
To assess global conformational changes brought about by ligand binding, we conducted nuclease V1 (helix-specific) and nuclease T2 (single-strand-specific) partial digests of the free and ligand-bound riboswitches. In the absence of TPP these nucleases cleave regions that participate in TPP binding, such as nucleotides (nt) 39–44 (J3-2) and nt 59–61 (part of P4/P5), as well as some regions that are not directly involved in formation of the TPP-binding pocket, such as nt 67–72 (L2) and nt 84–86 (Fig. 4 and Supplementary Figs S6 and S7). The addition of TPP decreases nuclease cleavage in regions that are important for both ligand binding and RNA folding (Fig. 4a, c). These results indicate that TPP not only stabilizes the two sections of the binding pocket by bridging the J3-2 and P4/P5 regions but also promotes the formation and/or stabilization of distal tertiary contacts within the J2-4/P2/P4 region and between L5 and P3.
Figure 4. Structural probing of the TPP riboswitch and implications for TPP-mediated gene repression.
a, Representative RNase V1 and T2 cleavage patterns for the thiM riboswitch (nt 1–166). 5′ 32P-labelled RNAs were treated by nucleases in the absence (−) or presence (+) of one, three and ten equivalents of TPP as described in Supplementary Methods and ref. 30. −OH and Bc stand for ladders prepared by partial digestion with alkali or B. cereus RNase, respectively. The major cleavage protections and enhancements in the presence of TPP are labelled with triangles and stars, respectively. b, Primer extension analysis in the absence and presence of TPP or TMP. RT pauses are indicated by arrows. c, Summary of structure probing experiments. Major TPP protections against V1 (red) and T2 (magenta) RNases are shown. The RT pauses are indicated by arrows. d, Typical mechanisms of TPP-specific gene repression. Top: translation initiation regulation (thiM genes). Bottom: transcription termination regulation (thiC genes). Complementary sequences and alternate base-pairing are shown in blue. SD sequence and initiation codon are shaded green. TPP and Mg2+ ions are depicted in red and magenta, respectively. e, Diagram of the OFF state of the E. coli thiM riboswitch.
Tertiary contacts associated with the TPP-stabilized fold of the riboswitch are also apparent in primer extension assays using reverse transcriptase (RT). In the presence of various analogues, including thiamine monophosphate (TMP), RT does not alter its pattern of pausing compared with that observed in the absence of ligand (Fig. 4b). This probably reflects the inability of TMP to span the two ligand-binding portions of the sensing domain fully and to stabilize the global fold, consistent with the progressive loss of binding affinity for thiamine ligands that carry one or no phosphates4. However, in the presence of TPP, RT pauses at two distinct positions (Fig. 4b). One pause at G86 occurs close to a pair of stacked tetrads anchoring tertiary contacts involving J2-4, P2 and P4, indicating that this conserved structural element becomes stabilized on ligand binding (Fig. 4c). A second RT pause occurs at C74 adjacent to the tertiary contact between L5 and P3, which seems to be sufficiently strong despite the disruption of base pairing in P1, P4 and P5 during primer extension.
The composite TPP-binding pocket can be defined as a molecular ruler in which only a ligand of proper length can stabilize tertiary interactions and modulate gene expression. This structural characteristic allows the riboswitch to achieve up to about 3,000-fold discrimination against shorter TMP and thiamine, and to provide feedback gene regulation only in the presence of biologically active TPP (ref. 4). In contrast to such unprecedented discrimination of related ligands by the RNA distance ruler, purine riboswitches have developed a completely different approach to distinguishing between related ligands. Adenine-specific and guanine-specific riboswitches use a single tight pocket to position their ligands precisely for Watson–Crick pairing with the discriminatory nucleotide, uracil or cytosine, respectively, thus making a single nucleotide the key element governing riboswitch regulation16,17.
Two predominant modes of TPP-dependent gene regulation, translation inhibition and transcription termination, are illustrated in Fig. 4d. Both require an over-threshold concentration of TPP in the cell for interaction with the riboswitch4,9,11,19. As evident from our biochemical experiments, binding of TPP to the riboswitch induces conformational rearrangements leading to stabilization of the overall RNA fold and, most importantly, to stabilization of conserved tertiary contacts adjacent to P1, as defined in the crystal structure (Fig. 4e). These interactions, in turn, stabilize P1 and promote folding of the expression platform, either to a hairpin that sequesters the Shine–Dalgarno (SD) sequence or to a terminator hairpin (OFF state); this results in the failure of translation initiation or premature transcription termination, respectively (Fig. 4d). Without TPP the riboswitch adopts alternative conformations, forming an anti-terminator hairpin or opening the SD sequence for ribosome binding (ON state).
Recent research19 has revealed that the antimicrobial compound PTPP (Fig. 1a) binds to bacterial and fungal TPP riboswitches and can turn off the expression of critical biosynthetic genes. The structure indicates that the loss of PTPP activity, associated with drug-resisting mutations in TPP riboswitches, might be due to the disruption of key tertiary contacts made by tetrads (C50G, A84G, G86A) and J3-2 elements (A47Δ) (Fig. 3a, b). PTPP carries a pyridine ring in place of the thiazole ring of TPP (Fig. 1a). Because the TPP riboswitch does not make any substantive contacts with the ligand in this chemically distinctive region, our structure-based model reveals why PTPP functions as a mimic of the natural ligand (Supplementary Fig. S8). Given the important functional role of riboswitches in numerous microorganisms and the fact that riboswitches have not yet been detected in the human genome, structures of TPP and other riboswitch classes should enable researchers to employ rational drug discovery strategies to create novel classes of antibacterial and antifungal compounds that target riboswitches19,26,27.
METHODS
Crystallization
The TPP–riboswitch complex was prepared by mixing RNA transcribed in vitro and TPP in a buffer containing 50 mM potassium acetate pH 6.9 and 5 mM MgCl2. Crystals were grown by hanging-drop vapour diffusion. The complex solution and reservoir solution (28% w/v poly(ethylene glycol) 4000, 100 mM sodium acetate pH 4.8 and 200 mM ammonium acetate) were mixed in a 1:1 ratio and incubated at 20°C. For soaking, crystals were washed with stabilizing solution lacking MgCl2 and then incubated in the presence of 3 mM Os(NH3)6 for two days.
Structure determination
Native and Os(NH3)6 multiple anomalous diffraction (MAD) data were collected at beamline X25 at the Brookhaven National Synchrotron Light Source. Data were processed with the HKL2000 suite of programs (HKL Research). The structure was determined by using MAD osmium data and SOLVE/RESOLVE28. The RNA model was built using TURBO-FRODO (〈http://afmb.cnrs-mrs.fr/rubrique113.html〉) and refined with REFMAC29 using a native data set (Supplementary Figs S9–S11). TPP and cations were added to the model on the basis of analysis of 2F o − F c and F o − F c electron density maps. Na+, K+ and hydrated Mg2+ were modelled on the basis of the number of coordination bonds, their distances and their coordination geometry. RNA residue 55 has a partial electron density.
Supplementary Material
Acknowledgments
We thank L. Malinina and Y.-R. Yuan for extensive discussions; A. Kazantsev and M. Yusupov for the gift of Os(NH3)6; and M. Becker for assistance with synchrotron data collection on beamline X25 at the Brookhaven National Laboratory, supported by the US Department of Energy. D.J.P. and R.R.B. were supported by funds from the NIH.
Footnotes
Author Contributions A.S. was responsible for all structural and biochemical aspects of this project. A.P. participated in RNA preparation and crystallization trials, and A.T.P. participated in NMR experiments. D.J.P., A.S. and R.R.B. co-wrote the paper. All authors discussed the results and commented on the manuscript.
Author Information Coordinates of the X-ray structure of the TPP-bound riboswitch have been deposited in the RCSB Protein Data Bank under accession code 2GDI. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Mandal M, Breaker RR. Gene regulation by riboswitches. Nature Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 2.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Soukup GA, Soukup JK. Riboswitches exert genetic control through metabolite-induced conformational change. Curr Opin Struct Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 5.Schowen RL. In: Comprehensive Biological Catalysis. Sinnott M, editor. Vol. 2. Academic; San Diego: 1998. pp. 217–266. [Google Scholar]
- 6.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubodera T, et al. Thiamine-regulated gene expression of Aspergillius oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 8.Miranda-Rios J, Navarro M, Soberón MA. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci USA. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in prokaryotes. New genes and regulatory mechanisms. J Biol Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 10.Nahvi A, et al. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 11.Mironov AS, et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 12.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 14.Winkler WC. Metabolic monitoring by bacterial mRNAs. Arch Microbiol. 2005;183:151–159. doi: 10.1007/s00203-005-0758-9. [DOI] [PubMed] [Google Scholar]
- 15.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 16.Batey RB, Gilbert SD, Montagne RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 17.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auffinger P, Bielecki L, Westhof E. Anion binding to nucleic acids. Structure. 2004;12:379–388. doi: 10.1016/j.str.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Nagaswamy U, Fox GE. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA. 2002;8:1112–1119. doi: 10.1017/s135583820202006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamauchi T, et al. Roles of Mg2+ in TPP-dependent riboswitch. FEBS Lett. 2005;579:2583–2588. doi: 10.1016/j.febslet.2005.03.074. [DOI] [PubMed] [Google Scholar]
- 22.Brodersen DE, et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 23.Schlunzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 24.Pioletti M, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hesselberth JR, Ellington AD. A (ribo) switch in the paradigms of genetic regulation. Nature Struct Biol. 2002;9:891–893. doi: 10.1038/nsb1202-891. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–245. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Serganov A, Polonskaia A, Ehresmann B, Ehresmann C, Patel DJ. Ribosomal protein S15 represses its own translation via adaptation of an rRNA-like fold within its mRNA. EMBO J. 2003;22:1898–1908. doi: 10.1093/emboj/cdg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.