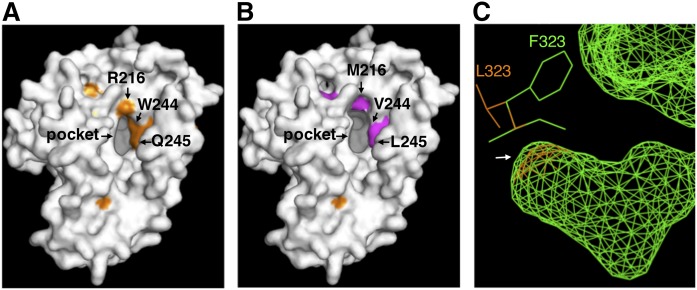
Fig. 7.
Homology modeling of three-dimensional structures of rat D6d (A) and the mutants D6d-mut8 (B) and D6d-L323F (C). The models were generated by using the protein structure prediction program Phyre2, with the crystal structure of human stearoyl-CoA desaturase as a template. Amino acid residues that were identified as determinants for substrate specificity and that are located around the entrance of the substrate-binding pocket are indicated by arrows. C: Compares the predicted structures of the bottom of the internal substrate-binding cavity (an arrow) and the vicinal amino acid residues of intact D6d (orange) and D6d-L323F (green).