Abstract
Streptococcus pyogenes chromosomal island M1 (SpyCIM1) integrates by site-specific recombination into the 5’ end of DNA mismatch repair (MMR) gene mutL in strain SF370SmR, blocking transcription of it and the downstream operon genes. During exponential growth, SpyCIM1 excises from the chromosome and replicates as an episome, restoring mutL transcription. This process is reversed in stationary phase with SpyCIM1 re-integrating into mutL, returning the cells to a mutator phenotype. Here we show that elimination of SpyCIM1 relieves this mutator phenotype. The downstream MMR operon genes, multidrug efflux pump lmrP, Holliday junction resolution helicase ruvA, and DNA base excision repair glycosylase tag, are also restored to constitutive expression by elimination of SpyCIM1. The presence of SpyCIM1 alters global transcription patterns in SF370SmR. RNA sequencing (RNA-Seq) demonstrated that loss of SpyCIM1 in the SpyCIM1 deletion mutant, CEM1Δ4, impacted the expression of over 100 genes involved in virulence and metabolism both in early exponential phase, when the SpyCIM1 is episomal, as well as at the onset of stationary phase, when SpyCIM1 has reintegrated into mutL. Among these changes, the up-regulation of the genes for the antiphagocytic M protein (emm1), streptolysin O (slo), capsule operon (hasABC), and streptococcal pyrogenic exotoxin (speB), are particularly notable. The expression pattern of the MMR operon confirmed our earlier observations that these genes are transcribed in early exponential phase but silenced as stationary phase is approached. Thus, the direct role of SpyCIM1 in causing the mutator phenotype is confirmed, and further, its influence upon the biology of S. pyogenes was found to impact multiple genes in addition to the MMR operon, which is a novel function for a mobile genetic element. We suggest that such chromosomal islands are a remarkable evolutionary adaptation to promote the survival of its S. pyogenes host cell in changing environments.
Introduction
Prophages and prophage-like elements are universal components of the genomes of Streptococcus pyogenes (group A streptococcus), with the published genome sequences having between two and eight examples in each strain [1–12]. These important mobile genetic elements (MGE) provide a substantial contribution of genetic material to their hosts, often representing ~10% of the total genome. Streptococcal prophages generally follow the typical genetic organization of lambdoid phages with genes organized for coordinated expression of the establishment of lysogeny or for the expression of early and late genes in the lytic cycle [13, 14]. Importantly, superantigens and other streptococcal virulence factors are components of these prophage genomes. In addition to typical prophages, other MGEs are present in the S. pyogenes genomes that include insertion sequence (IS) elements, transposons, and chromosomal islands.
Recently, we demonstrated that a prophage-like MGE in the S. pyogenes M1 genome strain SF370 acted as a genetic switch that controlled the expression of the DNA mismatch repair (MMR) gene mutL as well as additional downstream genes. These genes are encoded on a polycistronic mRNA along with mutS, and the result of this regulation caused a growth-dependent mutator phenotype [15]. This MGE, which was originally annotated in the genome as prophage SF370.4 and is now named SpyCIM1 (S treptococcus py ogenes chromosomal island M1), mediated expression of the MMR operon through a process of dynamic excision and re-integration from the 5’ end of the mutL ORF. During a state of rapid cell division, the prophage element excises from the bacterial chromosome and replicates as a circular episome, allowing the normal expression of mutL and the downstream genes. When the cells approach stationary phase and division slows, the episomal form re-integrates into its attachment site in mutL, silencing the expression of mutL and downstream genes. This cycle of excision and re-integration results in the cell switching between a complex mutator and normal phenotype [15]. This system is remarkable in that not only is MMR regulated by this MGE, but it also controls a multiple drug efflux pump (lmrP), a Holliday junction helicase subunit (ruvA), and a component of base excision repair (tag). We extended this original observation to demonstrate that the frequent carriage of related SpyCI elements in the genomes of other S. pyogenes strains also was associated with a mutator phenotype [16].
The prophage-like element SpyCIM1 differs from typical integrated streptococcal prophages in a number of characteristics. Genetic modules for lysogeny, regulation, and DNA replication are readily identified by homology to these regions in other prophages, but no capsid or packaging structural genes, lysis genes, or virulence genes seem to be present. Further, SpyCIM1 and related elements found in other S. pyogenes genomes are smaller than typical streptococcal prophages, having a range between 13 kb to 17 kb in length [15, 16]. At first glance, such an element might be classified as a defective prophage as indeed it originally was [1, 14]. However, SpyCIM1 is a member of a sizeable group of elements in Gram-positives that follow a similar genetic organization, and it is unlikely that each chromosomal island resulted from independent prophage decay in these different genera of bacteria. So, as proposed by Novick and colleagues [17, 18], these phage-like chromosomal islands probably represent a separate class of mobile genetic elements.
Our previous studies showed that strains with SpyCI had a higher mutation rate and other phenotypic changes compared to strains lacking these MGEs; however, these earlier studies were limited to comparing S. pyogenes SpyCI+ strains to similar but genetically distinct ones lacking a SpyCI integrated into mutL [16]. Thus, the association between SpyCI carriage and a mutator phenotype was inferential and not proven due to the lack of isogenic strains for this MGE. In this report we demonstrate that the removal of SpyCIM1 from strain SF370SmR relieves the mutator phenotype by decreasing the mutation rate as well as increasing resistance to ethidium bromide, UV irradiation and ethyl methanesulfonate, which result from the restoration of constitutive expression of mutL, lmrP, ruvA, and tag. Further, loss of SpyCIM1 also altered global gene expression patterns, which were unique to growth phases of the cell. These studies confirm the impact this phage-like chromosomal island has upon its host and will be the springboard for future studies about this remarkable genetic system.
Materials and Methods
Strains and bacterial growth conditions
Bacterial strains and plasmids used in these studies are described in Table 1. Escherichia coli, used for vector propagation, was grown in Luria-Bertani (LB) broth (Difco) containing 50 μg/ml kanamycin sulfate (Amresco, Solon, OH). S. pyogenes strains were grown in Bacto Todd Hewitt Broth (Becton, Dickinson and Company, Sparks, MD) supplemented with 2% Bacto yeast extract (THY medium) at 37°C or Brain Heart Infusion media (BHI) (Himedia Laboratories, India) at 37°C with the addition of the appropriate antibiotics (Sigma); growth was determined by monitoring the absorbance at 600 nm. Strain SF370SmR is a streptomycin resistant derivative of strain SF370, containing a spontaneous mutation in the rpsL gene [19]. No differences between SF370 and the streptomycin resistant derivative were observed with respect to growth rate or in the various biological assays employed [19]. Chemically defined media (CDM) was prepared as described in the literature [20].
Table 1. Strains, plasmid, and oligonucleotide primers used in this work.
| Strains | Genotype/Relevant characteristics | Reference |
| SF370 | emm1 (contains prophages ϕSF370.1, ϕSF370.2, ϕSF370.3, and SpyCIM1) | [1, 14, 15] |
| SF370SmR | Spontaneous streptomycin resistant derivative of SF370 | [19] |
| CEM1KRΔ Spy2136 | SF370SmR ΔSpy2136::(aacA-aphD) / (rpsL WT )) | [19] |
| CEM1Δ4 | SF370SmRΔSpyCIM1; cured of SpyCIM1 | [19] |
| MGAS5005 | emm1 (SpyCI-free M1 strain) | [21] |
| K56 | emm12 (SpyCI-free M12 strain) | [22] |
| Plasmids | Description | |
| pFWKR | Counter-selection vector with janus cassette, (aacA-aphD) / (rpsL WT )) between MCS for phage elimination | [19] |
| pFWKR- Spy2136 | pFWKR with flanking regions of Spy2136 for inactivation of SpyCIM1 primase gene | [19] |
| PCR Primers | Sequence | Product size |
| mutSL-L | 5' AATCGCCAGT TCCTGATGTC | 1361 bp |
| mutSL-R | 5' GGGCTGCTGA TGATTTGATT | |
| int4-L | 5’ GTCGCTGTCT CATTTGATAG AGCTT | 401 bp |
| int4-R | 5’ CCAACAAGGA GTATTGCTAG GGC | |
| qRT-PCR Primers | Sequence | Product size |
| nga_RT-L | 5' ACGTTAGCCG CAAATACCAC | 89 bp |
| nga_RT-R | 5' GCTTGTAACG TGGGAAGCTC | |
| slo-RT-F | 5' AAGCTCCGCC ACTCTTTGTGA | 103 bp |
| slo-RT-R | 5' GCACTAAAGG CCGCTTCAAC 3' | |
| norA_RT-L | 5' AAACGACGAC CAAACACCTC | 102 bp |
| norA_RT-R | 5' TCAGACTAGC CAGGCAGGAT | |
| emm RT-L | 5' GCAAAACTAA GAGCTGGAAA | 139 bp |
| emm RT-R | 5' TAGTTTCCTT CATTGGTGCT | |
| speB RT-L | 5' AGCAGTTGCA GTAGCAACACAT | 141 bp |
| speB-RT-R | 5' CTCCTTGATT CAAAAGGCATTC | |
| hasB_RT-L | 5' TCCTCAAACG CTAATTGAAGC | 92 bp |
| hasB_RT-R | 5' CCCGCTCTTC TAAGACGTTG | |
| 16S_rRNA-L | 5’ AGCGTTGTCC GGATTTATTG | 126 bp |
| 16S_rRNA-R | 5’ CACTCTCCCC TTCTGCACTC |
Elimination of SpyCIM1 from S. pyogenes SF370SmR
A two-step phage counter-selection method using a Janus cassette was used to derive isolates that lost SpyCIM1 from the genome of SF370SmR; the complete details are described in Euler et al. [19]. In this strategy, the initial introduction of the Janus cassette results in the loss of streptomycin resistance and the acquisition of kanamycin resistance through replacement of the SpyCIM1 primase gene. Loss of the primase leads to instability of SpyCIM1 following excision, which promotes loss of this element. Curing of SpyCIM1 is then detected by a loss of kanamycin resistance with a concomitant restoration of streptomycin resistance. The SmR / KanS mutants identified following this process were then verified for the complete loss of the SpyCIM1 by PCR (using primers mutSL-L and mutSL-R to amplify the SpyCIM1 attachment site), Southern blot hybridization analysis (with SpyCIM1 specific gene probes), DNA sequence analysis, and PFGE analysis [19]. The resulting mutant strain, CEM1Δ4, no longer contained the integrated SpyCIM1 DNA in the streptococcal chromosome; further, the loss of SpyCIM1 restored the precise mutS-mutL junction seen in SpyCI-free strains of S. pyogenes.
Chromosomal DNA Isolation, Sanger sequencing, and analysis
Isolation of streptococcal DNA was performed as previously reported [15, 23]. For isolation of stationary phase genomic DNA, a single colony of each S. pyogenes strain was used to inoculate 5 ml of fresh THY or BHI broth, which were allowed to grow >16 hours at 37°C in 5% CO2 before DNA isolation unless stated otherwise. Automated DNA sequencing was performed at the University of Oklahoma Health Sciences Center Laboratory for Genomics and Bioinformatics. Prior to sequencing, PCR products were treated with shrimp alkaline phosphatase and exonuclease I by incubation at 37°C for 60 min, followed by inactivation of the enzymes by heating at 85°C for 15 min or cleaned with the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI). Sequencing was performed using the same primers that were used for PCR. PCR and Quantitative real-time PCR (qRT-PCR) were done as previously described [15, 16], using the primers listed in Table 1.
Southern blot analysis
DNA was isolated from strains SF370SmR, CEM1Δ4, MGAS5005, and K56. Following digestion with HindIII, the generated fragments were separated on a 0.8% agarose gel with 0.5X tris-borate-EDTA (TBE) buffer and transferred to nylon membrane using standard methods [24]. A probe covering the SpyCIM1-free junction between mutS and mutL in strain MGAS5005 was prepared by PCR amplification of this region using primers MutSL-L and MutSL-R and the DIG-[11]-dUTP PCR labeling kit from Roche Life Science (Indianapolis, IN), following the manufacturer’s recommended protocol. Hybridization, washing, and detection of the probe bound to the blot was done also following the recommended protocol.
Determination of the spontaneous mutation rate
A fluctuation test based on the Luria and Delbrück assay [25, 26] was used to determine the spontaneous mutation rates of S. pyogenes strains as previously described [15]. Briefly, THY broth was inoculated with an isolated colony of the strain to be tested and incubated overnight at 37°C. The overnight culture was diluted 1:104 into 41 ml fresh THY broth such that the final cell density is <1000 cells/ml. The culture was dispensed as 1 ml aliquots into 31 sterile culture tubes. The cultures were grown for 24 hours at 37°C to achieve maximum cell density. One culture was used to determine total CFU by serial dilutions plated on THY agar, while the remainder were each mixed with 3.0 ml melted 0.6% agar in water (45°C) and overlaid onto THY agar plates supplemented with 2 μg/ml ciprofloxacin. After the agar overlay hardened, the plates were incubated at 37°C for >48 hours to allow appearance of antibiotic resistant mutants. The mutation rate with confidence limits was calculated for each strain using the software package ft (P.D. Sniegowski, Univ. Pennsylvania [27]). The data collected was averaged to determine mutation rate in mutations/generation (μ). Each determination was done at least three times.
Sensitivity of strains to UV irradiation
The sensitivity of S. pyogenes strains to killing by UV irradiation from a calibrated 254 nm germicidal lamp (120 μW/cm2) was done as previously described [15]. Serial dilutions (ten-fold) of cells were exposed to UV light for up to two minutes in 30 second increments, in a darkened room to prevent photoreactivation, and were subsequently incubated overnight at 37°C to allow appearance of survivors. UV irradiation assays were conducted a minimum of three times per strain to confirm results.
Growth inhibition by ethidium bromide
The resistance of strain SF370SmR or CEM1Δ4 was done using a modification of a previously reported method [28]. An overnight culture of S. pyogenes grown in THY broth at 37°C was diluted in fresh media to an absorbance at 600 nm of ~0.05. Incubation was continued at 37°C until A600 nm = 0.3, and then the culture was diluted as before into fresh media containing 0, 1, 1.5, 2.5, or 5 μM ethidium bromide. The concentration of the ethidium bromide stock was determined using the extinction coefficient (ϵ) at 480 nm = 5680 M-1 cm-1 [29]. Each culture was grown as ten replicates (200 μl each) in a Bioscreen-C Automated Growth Curve Analysis System (Growth Curves US, Piscataway, NJ) at 37°C without shaking. The absorbance at 600 nm for each culture was measured every 15 minutes for a total of 24 hours. The 3 replicates of each condition where averaged and normalized to the cultures not treated with EtBr using the software package Graphpad Prism 4 (GraphPad Prism Software, La Jolla, CA).
Ethyl methanesulfonate (EMS) mutagenesis
EMS mutagenesis was done using a modification of a previously described method [30]. THY broth (5 ml) was inoculated with a single colony of SF370SmR or CEM1Δ4 and incubated overnight at 37°C. The overnight culture was diluted into fresh THY (1:20 dilution) and grown at 37°C for two hours. A portion (0.5 ml) was spread on a THY agar plate containing 2 μg/ml ciprofloxacin and allowed to dry for 30 minutes. The EMS stock solution (1.145 g/ml; Sigma-Aldrich, St. Louis, MO) was diluted into sterile water to give a final working solution of 1 μg/ml; 15 μl of this solution was spotted on a sterile paper disk and placed in the middle of the agar plate that had been spread with the S. pyogenes strain. For a control, a paper disk that been spotted with distilled water was placed on a parallel agar plate for each strain. After overnight incubation at 37°C, the appearance of ciprofloxacin resistant mutants were tabulated.
Global transcriptional analysis
Overnight cultures of SF370SmR and CEM1Δ4 were diluted 1:20 in filter sterilized (0.22μ) THY broth. The cultures were grown at 37°C or at 39°C to early log phase (A600 ~0.25) or to late log phase (A600 ~0.55). The cells were harvested by centrifugation, resuspended in 1 ml of RNALater (Ambion #AM7021, Life Technologies, Grand Island, NY, U.S.A.) and stored at 4°C. To process the cells for RNA extraction, the cells stored in RNALater were collected by centrifugation (8000xg for 5 min. at 4°C), and the pellets were resuspended in 200μL of RNA Storage Solution (1 mM Sodium Citrate, pH 6.4). The streptococci were lysed by incubation for 5 min at room temperature with PlyC Lysin (provided by V.A. Fischetti). All subsequent steps were performed at 4°C. RNA was extracted using Trizol Reagent (Life Technologies) and chloroform, using the manufacturer’s recommended protocol. The RNA pellets were dissolved in water (Molecular Grade Water, G-Biosciences #786–293, St. Louis, MO, U.S.A.) and quantified using a Nanodrop 1000 Spectrophotometer (Thermo Fisher Scientific Inc, Wilmington, DE, U.S.A.). The RNA samples were stored at -75°C until needed for further processing. Before RNA sequencing analysis, 16S and 23S ribosomal RNA was removed the samples by using a MicrobExpress kit (Ambion #AM1905) following the manufacturer’s protocol. The quality of the enriched mRNA samples was confirmed using an Agilent 2100 BioAnalyzer (Agilent Technologies, Inc., Santa Clara, CA, U.S.A.). The samples were converted to cDNA using Superscript II (Invitrogen, Carlsbad, CA) and random hexamer priming by following the manufacturer’s protocol. Library construction and whole transcriptome analysis was done at the Laboratory for Molecular Biology and Cytometry Research core facility at the University of Oklahoma Health Sciences Center using an Illumina MiSeq Next-Generation Sequencer and following the manufacturer’s recommended protocols (Illumina, Inc., San Diego, CA). Sequence reads were referenced to the S. pyogenes SF370 genome, and analysis was done using the software package GeneSifter (Geospiza, Inc. Seattle, WA). A likelihood ratio test was used to calculate the ratio of SF370SmR RNA to CEM1Δ4 RNA for each gene in the SF370 Genbank annotation. A Benjamini and Hochberg correction was applied to the data. The GEO accession for the datasets is record GSE75633. For mapping of mRNA and determination of polycistronic operons, the software package Rockhopper (http://cs.wellesley.edu/~btjaden/Rockhopper/) was used [31].
Results
Elimination of SpyCIM1 from the genome of strain SF370
Our previous studies showed that SpyCIM1 controls a growth-dependent mutator phenotype in the S. pyogenes M1 serotype SF370 through a dynamic process of excision and re-integration into a unique site in DNA MMR gene mutL (S1 Fig). These studies suggested that having SpyCIM1 was associated with an increased mutation rate as well as sensitivity to lipophilic antimicrobials, UV irradiation, and DNA methylating agents [15, 16]. To confirm these results we chose to eliminate this chromosomal island and create an isogenic strain that would be wild type for the MMR operon. A two-step phage counter selection method was developed to derive isolates that had lost the 13.5 Kbp SpyCIM1 from the genome of SF370; this process has been described in detail elsewhere [19]. The initial step in the elimination process was the selection for a spontaneous streptomycin resistant derivative of SF370. This strain, SF370SmR, was used for all subsequent studies that compared it to the isogenic mutant, CEM1Δ4, cured of SpyCIM1. As shown in Fig 1, the loss of SpyCIM1 from the MMR operon could be confirmed by PCR and Southern blot. DNA was isolated from cells after overnight incubation at 37°C, a condition where we previously showed that SpyCIM1 would be integrated into the bacterial chromosome at attB [15]. The SpyCIM1 bacterial attachment site (attB) is the first 16 bases of the mutL ORF and begins 131 bases downstream of the mutS ORF. Under typical PCR conditions, this region can only be amplified if the chromosomal island is absent from the genome during logarithmic growth, when SpyCIM1 excises and replicates as an episome [15]. However, during stationary phase of growth the presence of the 13.5 Kbp of SpyCIM1 in the SF370SmR genome prevents normal amplification of this product by PCR. The removal of SpyCIM1 in the CEM1Δ4 mutant allowed for amplification of the mutS-mutL junction during both logarithmic and stationary phases of growth as occurs in SpyCIM1-free strains like MGAS5005 (Fig 1A). The loss of the chromosomal island also caused a change in the DNA hybridization pattern to this same region, with CEM1Δ4 having the same pattern as SpyCIM1-free strains (Fig 1B). The elimination of SpyCIM1 from the chromosome and the restoration of the normal DNA sequence observed in CEM1Δ4 was confirmed by DNA sequencing (not shown). Also, no SpyCIM1 genes could be amplified by PCR in strain CEM1Δ4, confirming that the element is lost and not translocated to a new site on the SF370SmR chromosome (not shown). Further, PFGE analysis of CEM1Δ4 digested genomic DNA showed no aberrant genomic DNA rearrangements were caused by the loss of SpyCIM1 [19].
Fig 1. Elimination of SpyCIM1 (ϕ370.4) from its host strain.
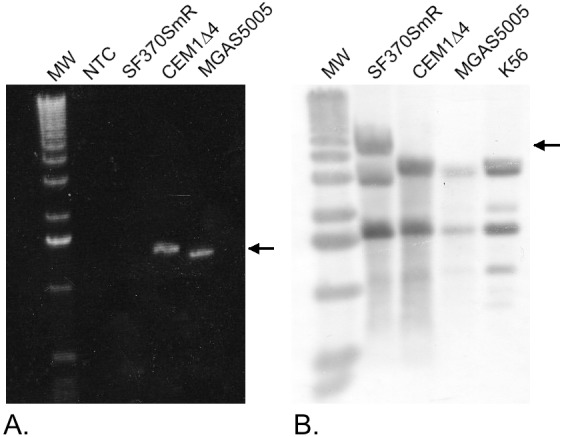
A. The prophage-free attB region can be amplified from a SpyCI-free M1 strain MGAS5005 and from SpyCIM1 cured strain CEM1Δ4, but not from the parental SF370SmR strain, where this region is interrupted by the presence of the 13.5 kb chromosomal island in the stationary phase cells. The arrow indicates the size of the expected 1361 bp PCR product following the loss of SpyCIM1. Lane 1 is a no template PCR control. B. Probing of a Southern blot of HindIII digested chromosomal DNA with a mutS-mutL probe showed SF370SmR has an altered pattern due to the presence of the integrated SpyCIM1 chromosome (indicated by the arrow), compared to CEM1Δ4, which has an identical hybridization pattern with SpyCIM1-free strains MGAS5005 and K56. MW: Kilobase molecular weight marker (Life Technologies, Grand Island, NY).
Elimination of SpyCIM1 reverses the mutator phenotype
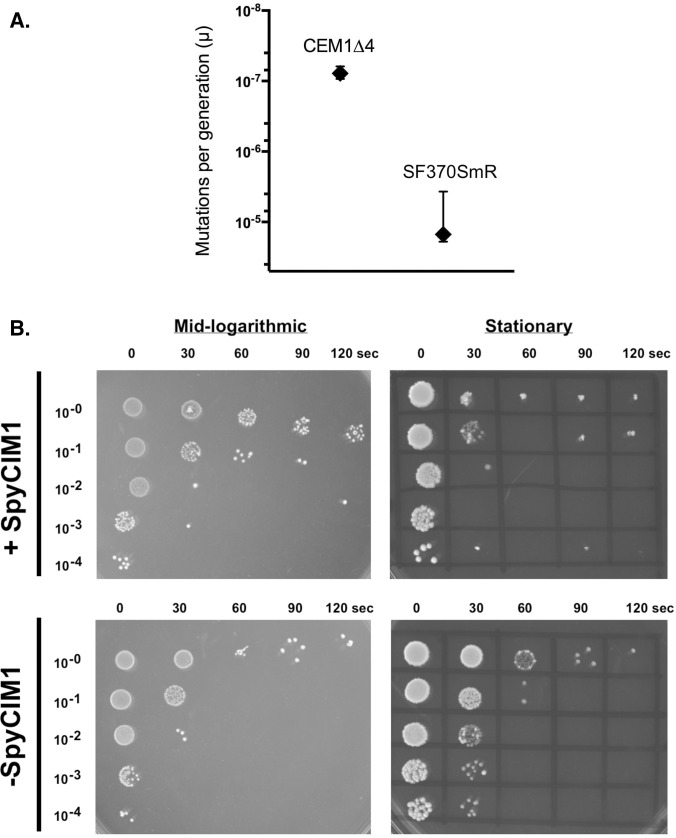
We previously demonstrated that the presence of SpyCIM1 correlated with a mutator phenotype that was evidenced by an increase in the spontaneous mutation rate and an increase in sensitivity to UV irradiation, due to the inhibition of mutL and ruvA expression, respectively [15]. For these studies, the appearance of spontaneous resistance to ciprofloxacin was used to determine these rates since resistance results from a limited number of specific single nucleotide changes in gyrA or parC [32, 33] and thus provides a good readout for observing spontaneous point mutations. As shown in Fig 2, the elimination of SpyCIM1 from SF370SmR caused a reversal of both mutator phenotypes. Using a modified Luria and Delbrück fluctuation test, the spontaneous mutation rate (mutations per generation) for parental strain SF370SmR was determined to be 1.6 X10-5, a value similar to our previous estimates [15, 16]. Elimination of SpyCIM1 resulted in a 200-fold decrease in the mutation rate (Fig 2A), similar to S. pyogenes strains lacking SpyCIM1, which ranged from 10−9 to 10−10 mutations per generation in our previous studies [15, 16].
Fig 2. A. The loss of SPyCIM1 reverses of the mutator phenotype in SF370SmR. The rate of spontaneous mutation to ciprofloxacin resistance for isogenic strains CEM1Δ4 (SpyCIM1-) and SF370SmR (SpyCIM1+) was determined by fluctuation test, and the rate of mutations per generation (μ) was calculated. For each strain, 30 parallel cultures were established with ~1,000 CFU/culture, grown for 24 h at 37°C, and plated individually on selective media. After 48 to 96 h of incubation, colonies were enumerated, and mutation rates with 95% confidence limits were calculated using the maximum likelihood estimation technique as implemented by the program ft [27, 34, 35]. SpyCIM1-free strain CEM1Δ4 showed a 200-fold reduction in the rate of spontaneous mutation as compared to SF370SmR. B. Enhanced resistance to UV irradiation following loss of SpyCIM1.
Ten-fold dilutions of strains SF370SmR and CEM1Δ4, which were growing in either logarithmic or stationary phase, were spotted onto an agar plate and exposed to 254 nm UV light (120 μW/cm2) for 0 to 120 sec. At logarithmic phase, when SpyCIM1 is excised from the chromosome [15], there was little difference in UV sensitivity between the strains. However, at stationary phase when SpyCIM1 is integrated into mutL, SpyCIM1 containing strain SF370SmR showed >100-fold more killing than isogenic strain CEM1Δ4, which lacks this element and is consistent with the restoration of ruvA expression. The protocol was performed in a darkened room to prevent photoreactivation. In the figure, the time in seconds (0–120) and the dilution factor of the cells are shown on the x-axis and y-axis, respectively.
In similar fashion, the loss of SpyCIM1 resulted in an increased resistance to killing by UV irradiation, reflecting the restoration of ruvA expression (Fig 2B). Comparing the survival of CEM1Δ4 to SF370SmR shows that SF370SmR has between 102 and 103 fewer survivors in the stationary phase for a given irradiation time. Interestingly, sporadic survivors were seen in SF370SmR at higher cell dilutions but not in CEM1Δ4 following longer irradiation times (>1 min), perhaps resulting from the mutator phenotype of strain SF370SmR (Fig 2A). The appearance of these isolated survivors in only the SpyCIM1+ strain, SF370SmR, was observed in every replicate of this experiment (not shown). By contrast, no clear difference is seen between the two strains at mid-logarithmic growth when SpyCIM1 is extrachromosomal. Interestingly, mid-logarithmic SF370SmR cells appear more resistant to UV irradiation at the longer exposures, a phenomenon that may be related to the global transcriptional changes described below. However, at stationary phase the loss of ruvA function appears dominant and clearly differentiates the phenotype of the two strains.
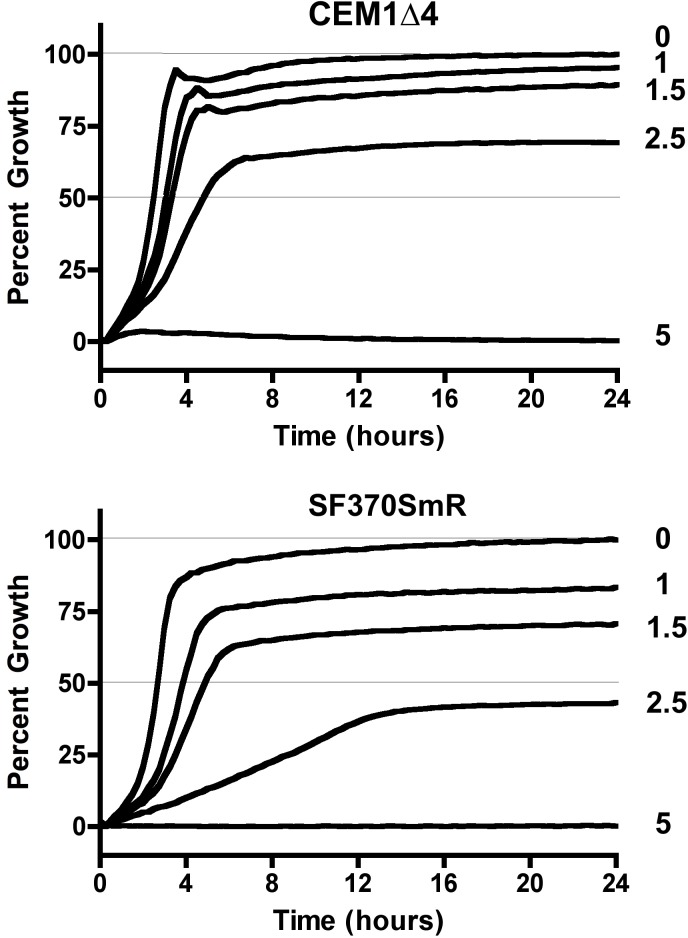
The lmrP gene is a member of the operon interrupted by the presence of SpyCIM1. The encoded protein is a member of the major facilitator superfamily (MFS) of drug efflux proteins and has a hydropathy profile that predicts LmrP to be an integral membrane protein with 12 transmembrane regions [36]. A homolog of this gene is found in Lactococcus lactis, providing resistance to ethidium bromide and other antimicrobial compounds [37, 38]. Strains SF370SmR and CEM1Δ4 were grown in the presence of increasing concentrations of EtBr (0 to 5 μM) over a 24-hour period, and cell growth was monitored by the absorbance of the culture at 600 nm. In the absence of EtBr, both strains achieved >85% of the culture’s maximum density after 4 hours (Fig 3) and similar absolute culture densities (not shown). Similarly, neither strain grew appreciably at concentrations of 5 μM EtBr. In the SpyCIM1-free strain CEM1Δ4, the addition of EtBr up to 1.5 μM reduced the final density of the culture only by 10%, and while 2.5 μM EtBr resulted in a substantial reduction in growth, the culture still achieved a density of 70% of the untreated cells. By contrast, when compared to CEM1Δ4, strain SF370SmR showed a marked increase in sensitivity to EtBr. Treatment with 1.0 μM resulted in nearly 20% growth reduction, and cultures treated with 2.5 μM achieved a 60% growth reduction compared to untreated cells. Furthermore, the rate of growth of SF370SmR was markedly slowed under these conditions, requiring over 12 hours to achieve maximum density. Thus, the removal of SpyCIM1 from SF370SmR to allow constitutive expression of lmrP resulted in a marked increase in resistance to EtBr.
Fig 3. Resistance to ethidium bromide in S. pyogenes with and without SpyCIM1.
Strains SF370SmR (lower panel) and its SpyCIM1 cured derivative CEM1Δ4 (upper panel) were grown at 37°C in THY broth with increasing concentrations of ethidium bromide (0, 1, 1.5, 2.5, and 5 μM). Each culture was grown in ten replicates, and growth was monitored every 15 minutes for 24 hours by the absorbance of the cultures at 600 nm. The data is presented as percentage of maximum growth observed for each strain without EtBr. Error bars, which were uniformly very small, are not shown for clarity of presentation. The numbers by each line indicate the concentration (μM) of EtBr in each culture.
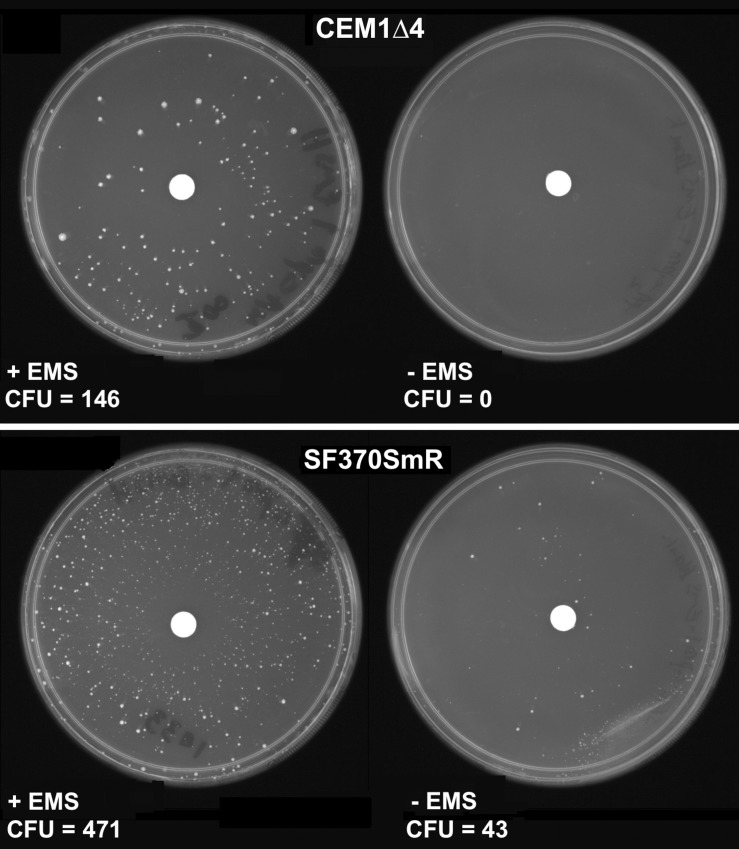
In E. coli, two 3-methyladenine-DNA glycosylases are present in the genome, encoded by alkA and tag [39]. These glycosylases are key components of base excision repair (BER), correcting a variety of alkylated bases that might lead to mutations; as seen with the mutagen EMS, which alkylates the O6 of guanine, leading to the formation O6-methylguanine causing G∙C → A∙T transitions [40, 41]. While no homolog has been identified in S. pyogenes to a known Gram-negative or -positive 3-meA DNA glycosylase II (alkA), a homolog of tag is located in the S. pyogenes MMR operon that would be controlled by SpyCIM1. Thus, the S. pyogenes tag gene product may play an essential role in the repair of alkylated bases, and its inhibition by SpyCIM1 could lead to an increased sensitivity to EMS or other DNA alkylating mutagens, resulting in an increased appearance of spontaneous mutations. Strains SF370SmR and CEM1Δ4 were treated with EMS to observe the appearance of chemically induced mutations leading to ciprofloxacin resistance. As shown in Fig 4, elimination of SpyCIM1 results in ~3-fold decrease in the appearance of spontaneous ciprofloxacin resistance in strain CEM1Δ4 after EMS treatment. Remarkably, even in the absence of EMS, SF370SmR still produced a few dozen spontaneous resistant mutants while CEM1Δ4 produced none. These studies show the increased sensitivity to induced mutagenesis by alkylating agents as well as the general mutator phenotype conferred by the presence of SpyCIM1.
Fig 4. SpyCIM1 enhances the appearance of mutants following EMS treatment.
Strains SF370SmR and CEM1Δ4 were spread on media plates containing ciprofloxacin (3 μg/ml). A paper disk containing either 15 ng EMS or sterile water was placed in the center of the plate, which was then incubated overnight at 37°C. The appearance of ciprofloxacin resistant mutants was observed in strain SF370SmR, even in the absence of EMS treatment (bottom right); under the same conditions strain CEM1Δ4 produced no resistant mutants (top right). Both strains produced ciprofloxacin resistant mutants following EMS treatment, but SF370SmR (bottom left) produced >3X as many resistant colonies as did SpyCIM1-free CEM1Δ4 (top left).
SpyCIM1 alters global transcription patterns in S. pyogenes
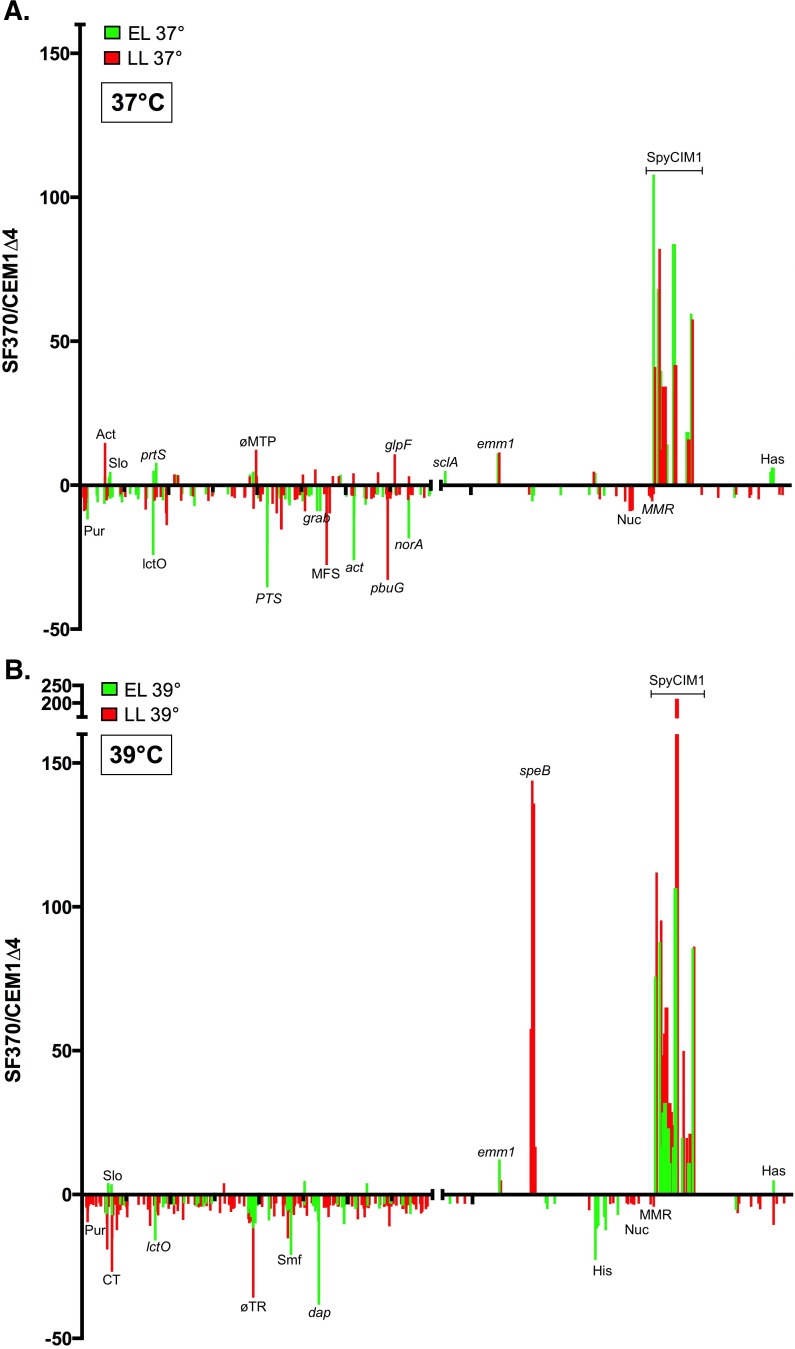
The studies presented here as well as our previous work argue that the expression of the MMR operon is controlled by the integrative state of SpyCIM1. Therefore, the global transcription patterns of SF370SmR and CEM1Δ4 were determined to observe whether the SpyCIM1 molecular switch might have an impact upon gene expression elsewhere in the S. pyogenes genome. Overnight growth of the two strains in CDM at 37°C suggested that their cell surface properties differed since SF370SmR had a “clumpy” phenotype (typical of high M protein surface expression) [42] while CEM1Δ4 did not (S2 Fig). RNA was isolated from SF370SmR and CEM1Δ4 at the initiation of logarithmic growth (early log phase; EL) and again late in logarithmic growth just before the cells entered stationary phase (late log phase; LL). High-throughput next generation RNA sequencing (RNA-Seq) was used to map and quantify the transcriptomes of these strains at those growth stages (Fig 5, Table 2, and S2–S6 Tables). The expression of the MMR operon was found to be the same in both strains at EL, in agreement with previous studies that demonstrated that SpyCIM1 excises from the chromosome and replicates as an episome at this time [15]. By LL the expression of the downstream MMR operon genes were depressed in SF370SmR by the re-integration of SpyCIM1, again in agreement with previous results. Analysis of the transcripts identified by RNA-seq allowed construction of a transcriptional map of SpyCIM1 (S4 Fig).
Fig 5. Elimination of SpyCIM1 alters global transcription patterns.
RNA was isolated from SF370SmR or CEM1Δ4 at the onset of logarithmic growth (Early Log) or immediately before the cells entered stationary phase (Late Log); the cells were either grown at 37°C (panel A) or 39°C (panel B). Samples were then analyzed by RNA sequencing (RNA-Seq). In the ratio of gene expression from SF370SmR compared to CEM1Δ4 for values greater than 3 or less than -3 are plotted against the corresponding gene identification number from the SF370SmR annotation [1]. The MMR operon showed that no difference in expression was observed between the strains during early log phase but that the transcription of these genes was inhibited in SF370SmR at late log phase, in agreement with previous studies [15]; an expanded view of this region is shown in S3 Fig. Notable genes whose expression was altered in SF370SmR or CEM1Δ4 are identified. The ticks on the X-axis mark every 500 genes. Legend: Act–acetate CoA-transferase operon; act–acetyltransferase Spy154; adhA–alcohol dehydrogenase; citM—Mg2+/citrate complex transporter; cspR - 23S rRNA methyltransferase; CT- conserved transmembrane protein Spy0169; dap—diaminopimelate epimerase; emm1 –M protein; glfP—glycerol uptake facilitator; grab—protein G-like alpha2-macroglobulin-binding protein; has–hyaluronic acid capsule operon; His–histidine catabolism operon; Hyp–gene encoding a protein of unknown function; lctO–lactate oxidase; MFS–uncharacterized major facilitator family protein (Spy1392); MMR–MMR operon; ϕMTP–phage major tail protein; norA—antibiotic resistance protein NorA; Nuc–nucleotide interconversions operon; pbuG—guanine-hypoxanthine permease; pncA—pyrazinamidase/nicotinamidase; prtS—cell envelope proteinase PrtS; PTS—mannose/fructose PTS system operon; pur–purine biosynthesis operon; sclA—collagen-like surface protein SclA; slo–streptolysin O operon; SpyCIM1 –phage-like CI. The identification of the SpyCIM1 genes is described in S1 Fig and S1 Table.
Table 2. Notable genes or operons whose transcription are altered by the presence of SpyCIM1.
The numbers in parenthesis are the fold induction or repression in transcription of strain SF370SmR as compared to CEM1Δ4. For operons, the induction or repression is the average fold change for the affected genes. A likelihood ratio test was used to calculate the ratio of SF370SmR RNA to CEM1Δ4 RNA for each gene in the SF370 Genbank annotation. A Benjamini and Hochberg correction was applied to the data using the software package GeneSifter. Ratios greater than 3 or less than -3 are reported. Data are taken from S2 Table.
| Early Logarithmic | Late Logarithmic | |||
| 37°C | 39°C | 37°C | 39°C | |
| Induced 1 | M-protein (+11.1) | M-protein (+11.4) | M-protein (+12.2) | M-protein (+5.0) |
| Capsule operon HasABC (+6) | Capsule operon HasABC (+3) | Major facilitator family protein NorA (+3.1) | Protease SpeB (+143.9) | |
| Collagen-like surface protein SclA (+4.9) | Transcriptional regulator Rgg3 (+3.8) | |||
| Streptolysin O operon (+3.0) | ||||
| Chemokine protease ScpC (C5a peptidase family) (+5.0) | ||||
| Early Logarithmic | Late Logarithmic | |||
| 37°C | 39°C | 37°C | 39°C | |
| Repressed | Arginine deiminase operon (-10.1) | Arginine deiminase operon (-4.9) | Purine biosynthesis operon (-7.0) | Purine biosynthesis operon (-4.7) |
| 67 kDa Myosin-crossreactive streptococcal antigen (-5.3) | 67 kDa Myosin-crossreactive streptococcal antigen (-3.2) | Guanine-hypoxanthine permease (-33) | Guanine-hypoxanthine permease (-11) | |
| L-lactate oxidase LctO (-24.3) | L-lactate oxidase LctO (-15.9) | Acetate CoA-transferase operon (-14.5) | Capsule operon HasABC (-6) | |
| Multidrug resistance protein NorA homolog (-18.5) | Multidrug resistance protein NorA homolog (-6.8) | Malic enzyme (MaeE) (-9.8) | Smf family DNA processing protein (-15) | |
| Thioesterase family protein Spy1339 (-9) | Thioesterase family protein Spy1339 (-38) | Nicotinate-nucleotide pyrophosphorylase (-12) | ||
| Transcriptional regulator RopB/Rgg1 (-3.6) | D-lactate dehydrogenase (-5.2) | Transcriptional regulator ComR (Rgg4) (-4.1) | ||
| Alcohol dehydrogenase (-11.9) | Histidine catabolism operon (-13) | Ferrichrome ABC transporter permease operon (-7.4) | ||
| ComYA (-5.9) | ||||
| Protein G-like alpha 2M-binding protein (GRAB) (-9) | ||||
| PTS system mannose/fructose family transporter operon (-18) | ||||
| Acetyltransferase Spy1546 (-26) | ||||
Surprisingly, the presence of SpyCIM1 correlates with global transcriptional changes in S. pyogenes with genes showing differential basal expression in EL, LL, or both (Fig 5A, Table 2, and S2 Table). While some of these global changes may be related to the differential expression of the MMR operon, other changes must map directly back to SpyCIM1 since in EL strains SF370SmR and CEM1Δ4 differ only by the presence or absence of the extrachromosomal form of this chromosomal island. Several key virulence genes have altered expression in SF370SmR; perhaps the most striking is the >11-fold higher expression of the major antiphagocytic M1 protein gene (emm1) in SF370SmR as compared to CEM1Δ4 in both EL and LL. The hyaluronic capsule operon (hasABC) is also more highly expressed in SF370SmR during EL as well as the virulence operon encoding NADase (nga) and streptolysin O (slo). Other SF370SmR genes were strongly depressed in EL as compared to CEM1Δ4, particularly the genes for the homolog of the MFS antibiotic resistance pump norA [43] and for lactate oxidase (lctO). The down-regulation of lctO may promote survival in SF370SmR by preventing lethal self-intoxication by H2O2 production [44]. Interestingly, norA, although depressed in EL, is more highly expressed in SF370SmR in LL. Inhibition of NAD-dependent malic enzyme MaeE has recently been linked to both S. pyogenes survival in low pH and to increased virulence [45], and this gene has nearly a 10-fold LL reduction in SF370SmR. Many changes related to metabolic pathways are also observed in the transcriptome of SF370SmR. The mannose/fructose phosphotransferase (PTS) operon (Spy1057-Spy1060) is down regulated in SF370SmR in EL, while an oxalate:formate antiporter (Spy1392), which is another MFS efflux protein, is strongly down regulated in LL (by ~27-fold). Many other metabolic changes are seen in SF370SmR, including depression of purine metabolism; the details are presented in S3 and S4 Tables.
Transcriptional patterns of cells grown at 39°C were examined also (Fig 5B, Table 2 and S2 Table). This temperature was chosen to mimic alterations in transcription that might result from growth in a febrile human. The M-protein (emm1) remained more highly expressed in SF370SmR regardless of growth phase or culture temperature. Similarly, the capsule operon (hasABC) remained more highly expressed in EL while L-lactate oxidase (lctO), the arginine deiminase operon, the multidrug resistance protein norA homolog, and the 67 kDa myosin-crossreactive streptococcal antigen were all decreased in expression in SF370SmR during EL at either 37° or 39°C. Other genes were altered in expression in a temperature dependent fashion, including several global transcriptional regulators. Transcriptional regulator Rgg3 has increased expression at 37°C during late log growth but not at 39°C while RopB (Rgg1) and ComR (Rgg4) are repressed by the presence SpyCIM1 in early log growth at 37°C and late log growth at 39°C, respectively. The most dramatic shift in expression, however, occurs in the gene for SpeB, which is increased over 140-fold in SF370SmR over CEM1Δ4 at 39°C in late log cells; qRT-PCR from the same cDNA preparations confirmed this difference in expression between the strains (S7 Table). To determine if this induction in speB expression was reproducible, three independent cultures of both strains were grown at 39°C, and samples were removed for RNA isolation at EL and LL as before. An additional sample was taken one hour after LL. The cDNA from these RNA samples were used as template for qRT-PCR to determine the expression of speB and other genes showing differential expression (S8 Table). In agreement with the RNA-seq data, this analysis confirmed the enhanced expression of speB as well as the differential expression of nga, slo, norA, emm, and hasB (S8 Table). Taken together, these results argue that SpyCIM1 encodes some mechanism that alters the transcription program of SF370SmR in a way that extends beyond only regulation of the MMR operon.
Discussion
In these studies, we demonstrate how the elimination of SpyCIM1 from the chromosome of S. pyogenes strain SF370SmR resulted in a decreased spontaneous mutation rate and increased resistance to ethidium bromide, UV irradiation, and EMS mutagenesis, by allowing constitutive expression of the MMR operon. Our previous studies relied on inference to demonstrate the impact of SpyCIM1 on the host phenotype through the comparison of unrelated streptococcal isolates [15, 16]. The present studies provide direct evidence for the SpyCIM1 associated mutator phenotype using isogenic strains that differed only by the presence of this chromosomal island. Further, these studies show that elimination of SpyCIM1 alters global transcription in the host streptococcus cell, and that these changes could potentially reduce virulence. Thus, the data provide important new information on the emerging field of Gram-positive phage-like chromosomal islands that alter the host bacterium’s phenotype.
The chromosomal island SpyCIM1 is a dynamically active phage-like element that alternates between chromosomal and episomal states, which mediates phenotypic changes upon the host bacterium. The penetrance of these phenotypes reflects the relative proportion of the population that has SpyCIM1 in the excised or integrated form. Therefore, an observed phenotype might vary over time as environmental or physiological conditions could favor the integrated or episomal state of SpyCIM1. An example of such changes was illustrated in Fig 4, where the resistance to UV irradiation was essentially the same in cells having or lacking SpyCIM1 during logarithmic growth when SpyCIM1 was episomal. However, a distinct phenotype became apparent when the cells were in stationary phase and SpyCIM1 was integrated into the bacterial chromosome. It is probable that SpyCIM1 mobilization during normal cell division results from an event that is linked to the cell cycle, but the process also appears to be inducible by DNA damage since mitomycin C treatment promotes SpyCIM1 excision [15]. Little is known about the details of SOS repair and its induction in S. pyogenes; indeed, no homolog of LexA is readily identifiable. Therefore, the molecular control of SpyCIM1 mobilization either triggered by SOS repair or by normal growth remains to be discovered.
Global transcriptional analysis showed the basal expression of numerous genes is altered by the presence of SpyCIM1. While notable differences could be seen between SF370SmR and CEM1Δ4 in both EL and LL cultures, the role of SpyCIM1 is clearly evident in the EL comparisons when the two strains differ only by the presence of the extrachromosomal chromosomal island in SF370SmR (Fig 5 and S2 Table). Thus, the transcriptional differences in EL do not result from transcriptional inhibition of the MMR operon but from the presence of SpyCIM1 encoded gene products in the cell.
The global gene expression changes associated with SpyCIM1 appear to either occur independently of target subsets of known S. pyogenes regulatory networks. For example, the Mga regulon includes a number of virulence genes such as the M protein (emm1), C5a peptidase (scpA), secreted inhibitor of complement (sic) and streptococcal collagen-like protein (scl1/sclA) [46]. In EL, the expression of the emm1 is 11-fold and sclA ~5-fold higher in SF370SmR, respectively, as compared to CEM1Δ4. By contrast, the C5a peptidase and Sic mRNAs are expressed equally in both strains, as is the expression of Mga itself. Therefore, the differences in gene expression seen between the two strains appears to step outside of the characterized regulatory networks in group A streptococci [46], suggesting that some SpyCIM1 encoded factor is responsible. SpyCIM1 contains a number of genes encoding proteins with a predicted helix-turn-helix (HTH) structure (S1 Table). The HTH products of genes Spy2125 and Spy2126 probably function as the repressor and antirepressor for SpyCIM1 integration and excision, given their position and orientation in the genome that is analogous to the lysogeny module of many prophages; however, there are other SpyCIM1 genes encoding HTH proteins (Spy2127, Spy2134, and Spy2145) whose biological role is currently unknown. Mga binding sites are reported to be diverse, having only 13.4% identity [47], and so some other DNA binding proteins such as the ones encoded by SpyCIM1 could potentially target only a sub-set of this regulon.
Many of these transcriptional changes may enhance virulence or survival. The increase in M protein and capsule expression would be antiphagocytic while the repression of arginine deiminase [48] and L-lactate oxidase [44, 49] at 37°C in EL may enhance virulence or prevent autointoxication by hydrogen peroxide production, respectively. The expression of the exotoxin SpeB is influenced by a number of S. pyogenes transcriptional regulators [50], and so, it is not surprising that it and its co-transcribed genes are stimulated in expression under some conditions (LL and 39°C), although the degree of induction seen was striking (>140-fold in SF370SmR as compared to CEM1Δ4; Table 2); qRT-PCR confirmed this difference in expression from identical samples between the strains (S7 Table). This induction is specific for 39°C since no differences in expression between the two strains was observed for SpeB at 37°C. Further, while the degree of induction was not as high as in the original experiment, growth of replicate cultures of both strains at 39°C confirmed the enhanced expression of speB (average fold induction in SF370SmR over CEM1Δ4 = 18.3 ± 11.9), and the differential expression of the other genes analyzed (S8 Table). Since it has been demonstrated previously that the induction of speB is sensitive to the growth state of the cell as well as environmental conditions [51–53], it is possible that SpyCIM1, either directly or indirectly, alters the timing of speB expression in LL at 39°C, leading to earlier expression of the gene in SF370SmR. Sampling of RNA from these cultures one hour later eliminates this difference, and indeed, CEM1Δ4 has about a ~2 fold greater expression of speB by this stage of growth (S8 Table). Future studies will be needed to clarify the differences in kinetics of speB expression between SF370SmR and CEM1Δ4 in fine detail.
In E. coli, it has been reported that select RNA chaperones and a cold shock protein are overexpressed to compensate for a mutator phenotype [54]. None of the potential DEAD box helicase RNA chaperones (Spy0288, Spy1369, Spy1659, Spy1837, and Spy1415) were altered in expression by the loss of SpyCIM1; however, cold-inducible RNA chaperone and antiterminator protein Csp (Spy2077) was inhibited 5.5-fold in SF370SmR at 39°C in LL. Thus, the inhibition of Csp may further enhance the SpyCIM1-associated mutator under these conditions. These shifts in gene expression, which appear to cut across a number of known regulatory networks, may improve fitness of the SpyCIM1 host cell. The potential benefits to a host bacterium following phage toxigenic conversion have been proposed many times [55–58], but the lack of such an identifiable virulence factor in SpyCIM1 led to the suggestion that it was a prophage remnant [1, 14]. While homologs of SpyCIM1 have been frequently observed in subsequent S. pyogenes genomes, their descriptions as a nonfunctional cryptic element remained until their role in regulating the MMR operon was discovered [15]. The results presented here argue that the impact of SpyCI on streptococcal survival and virulence extends well beyond controlling this operon.
Several qualifications must be considered in interpreting the transcriptional analysis data. The populations of cells are not synchronous so many differences seen may be the average expression of genes from cells that may be in different stages of growth; this situation is particularly important in SF370SmR where the population is undoubtedly a mixture of cells having the integrated or episomal form of SpyCIM1 [15]. Further, the altered expression of some genes may not be a direct result of some SpyCIM1-encoded gene product but result from the altered expression of some other host gene or regulatory network by the chromosomal island. Other differences in expression between cells grown at 37°C or 39°C may be due to enhanced or accelerated mRNA turnover at the higher temperature. Some changes, however, must map to altered promoter function caused either by a SpyCIM1 encoded product (protein or RNA) or perturbation of some S. pyogenes regulatory network.
Altered expression was observed in some genes encoding known regulatory proteins, which could contribute to the differing phenotypes. Comparing the expression of SF370SmR to CEM1Δ4, ropB (rgg1; Spy2042) was inhibited >3X at 37°C and EL, rgg3 (Spy0533 [20]) was increased >4X at 37°C and LL, and comR (rgg4; Spy0037) was inhibited >4X at 39°C and LL. Interestingly, the RNA-seq data showed that RofA-like gene ralp3 [59], which has a frameshift in SF370SmR, was expressed as a truncated transcript encoding amino acids 411 through 461 of the complete protein; the significance of this expression is unknown. Some of these shifts in regulation may influence horizontal transfer in group A streptococci. Although our understanding of group A streptococcal transformation is at an early stage [60], the inhibition of ComR (Rgg4) expression in SF370SmR at 39°C and LL might inhibit both competence and biofilm formation as recently demonstrated [60]. Perhaps related to controlling horizontal transfer, the Smf family DNA processing protein/DNA polymerase sliding clamp subunit (Spy1163) is also inhibited under the same conditions, and this protein functions as a transformation-dedicated DNA loader for RecA in S. pneumoniae [61]. Genes ComEC and ComEA, encoding homologs of the DNA translocation machinery channel protein in S. pneumoniae [62] are similarly inhibited by these conditions. Since Marks and coworkers found that group A streptococcal competence was highest at 34°C and decreased when the temperature was raised to 37°C [60], it is perhaps not surprising that a further increase to 39°C would down-regulate any associated genes. However, our transcriptional studies show that the presence of SpyCIM1 appears to enhance this limitation to horizontal transfer at elevated temperatures.
The regulation of the gene expression in Gram-positive bacteria by small, phage-like chromosomal islands is an emerging theme in prokaryotic biology [18, 63], and the examples that have been identified so far modify host cell function in a variety of ways from toxigenic conversion to gene regulation. We have shown here the direct impact SpyCIM1 has upon the expression of the MMR operon in S. pyogenes, but many questions concerning this system remain to be answered. For example, the activation of SpyCIM1 excision appears to be linked to the onset of cell division [15], but the cellular signals that trigger this event as well as the subsequent molecular decision to re-integrate into mutL remain unidentified. However, the most interesting field for exploration is how SpyCI promotes host fitness or survival, particularly in natural human infections. The acquisition of a mutator phenotype appears to increase bacterial virulence under some circumstances as a recent study of base excision repair mutants in Streptococcus mutans demonstrated [64]. The dynamic nature of this chromosomal island may prove to be an essential component of its relationship with S. pyogenes. Since MMR deficient strains are frequently found in many bacterial species, it might be inferred that inactivation of this repair system alone should not destabilize a cell. However, the one example we have of a defective and permanently integrated SpyCI (in the M5 genome strain Manfredo) did not result in silencing the MMR operon but rather demonstrated rescue of the operon through the evolution of a cryptic promoter within the chromosomal island remnant [16]. This result suggests that rounds of MMR operon expression and silencing may prove to have strong selective value, allowing a phenotypic flexibility that is unobtainable with simple gene inactivation through mutation. Thus, the dynamic control of this repair system by SpyCIM1 is an ideal method for achieving the mutator phenotype while minimizing the risks associated with long-term hypermutability.
In spite of decades of antibiotic therapy, S. pyogenes has shown a remarkable ability to maintain its niche as a major human pathogen; it has the ability to infect, colonize, and rapidly adapt to multiple environments in the human body with different tissue tropisms, while periodically producing novel strains associated with severe disease outbreaks. The novel, switchable mutator phenotype coupled with global transcriptional changes conferred by the SpyCI element may prove to be an important factor contributing to the adaptability and the evolutionary robustness of this pathogen.
Supporting Information
Phage-like chromosomal island SpyCIM1 regulates the expression of an operon containing genes mutL, lmrP, ruvA, and tag in response to cell growth [15, 16]. The presence of an integrated SpyCIM1 results in the cells adopting a mutator phenotype with regard to DNA mismatch repair (MMR), multiple drug efflux, Holliday junction resolution, and base excision repair. In early logarithmic phase, SpyCIM1 excises from the SF370 chromosome, restoring expression of mutL and the downstream genes (insert). As the cells approach stationary phase, SpyCIM1 re-integrates into mutL, silencing gene expression. This process of SpyCIM1 excision and integration causes the cells to alternate between a wild type and mutator phenotype. In the figure, transcriptionally active MMR operon genes are black while inactive ones are gray. The identification and potential function of the SpyCIM1 genes are detailed in S1 Table). Adapted from Nguyen and Scott [63], and used by permission.
(TIF)
Strains SF370SmR and CEM1Δ4 were grown overnight at 37°C in CDM. Gentle shaking was used to simultaneously disperse each cell pellet, showing that SF370SmR had a clumping phenotype that CEM1Δ4 lacked. The cultures were streaked on blood agar to confirm that both were pure cultures of each starting strain.
(TIF)
The data are taken from Fig 5. The first gene in the MMR operon, mutS, is not shown since no differences in transcription were ever observed between SF370SmR and CEM1Δ4. Only differences in transcription ≥ ±3-fold are shown.
(TIF)
Transcriptome analysis of mRNA from SF370SmR allowed prediction of the messages encoded by SpyCIM1, including probable polycistronic ones. The genetic map of SpyCIM1 is shown flanked by the MMR operon genes. Row A below the genetic map shows the predicted mRNAs that match genes in the SF370SmR annotation. Row B shows small RNAs that were detected by RNA-Seq but which are not included in the Genbank annotation. The small RNA immediately upstream of the int mRNA encodes a small peptide with a transmembrane domain. Prediction was accomplished using the software package Rockhopper [31].
(TIF)
The protein IDs refer to the ORF numbers in the published SF370 genome [1]. Products are predicted by BLASTP homology of the encoded proteins to GenBank entries. Several additional ORFs have been identified since the original annotation was prepared.
(PDF)
A likelihood ratio test was used to calculate the ratio of SF370SmR RNA to CEM1Δ4 RNA for each gene in the SF370 Genbank annotation. A Benjamini and Hochberg correction was applied to the data using the software package GeneSifter. Ratios greater than 3 or less than -3 are reported and were used to create the plot in Fig 5.
(PDF)
The analysis was done using GeneSifter as above.
(PDF)
The analysis was done using GeneSifter as above.
(PDF)
The analysis was done using GeneSifter as above.
(PDF)
The analysis was done using GeneSifter as above.
(PDF)
The cDNA preparations used for RNA-seq analysis were analyzed by qRT-PCR for comparing the expression of nga, slo, norA, emm1, speB, and hasB in SF370SmR to CEM1Δ4 as described in the Methods.
(PDF)
Three independent cultures of SF370SmR and of CEM1Δ4 were grown at 39°C; samples were harvested for RNA isolation when the culture density (A600 nm) was 0.2 (EL) and again when the density was 0.5 (LL). An addition sample was harvested one-hour post LL (Stationary). After conversion of the RNA to cDNA, qRT-PCR was used to compare expression levels of the listed genes between SF370SmR and CEM1Δ4. Values are the average and standard deviation of the fold-difference between the two stains.
(PDF)
Acknowledgments
This work was made possible by an Oklahoma Center for the Advancement of Science and Technology (OCAST) grant HR11-133 and by NIH Grant Number R15A1072718 to WMM and NIH Grant AI11822 to VAF.
Data Availability
Data are available in the GEO database, accession record GSE75633 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75633).
Funding Statement
NIH Grant R15A1072718 www.nih.gov WMM, NIH Grant AI11822 www.nih.gov VAF, and OCAST grant HR11-133 www.ok.gov/ocast/ WMM.
References
- 1. Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, et al. Complete Genome Sequence of an M1 Strain of Streptococcus pyogenes . Proceedings of the National Academy of Sciences of the United States of America. 2001;98(8):4658–63. 10.1073/pnas.071559398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, et al. Evolutionary Origin and Emergence of a Highly Successful Clone of Serotype M1 Group A Streptococcus Involved Multiple Horizontal Gene Transfer Events. J Infect Dis. 2005;192(5):771–82. . [DOI] [PubMed] [Google Scholar]
- 3. Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, et al. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(15):10078–83. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakagawa I, Kurokawa K, Yamashita A, Nakata M, Tomiyasu Y, Okahashi N, et al. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 2003;13(6A):1042–55. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holden MT, Scott A, Cherevach I, Chillingworth T, Churcher C, Cronin A, et al. Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain manfredo. Journal of bacteriology. 2007;189(4):1473–7. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banks DJ, Porcella SF, Barbian KD, Beres SB, Philips LE, Voyich JM, et al. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. Journal of Infectious Diseases. 2004;190(4):727–38. . [DOI] [PubMed] [Google Scholar]
- 7. Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, Deleo FR, et al. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus . Proceedings of the National Academy of Sciences of the United States of America. 2006;103(18):7059–64. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, Sylva GL, et al. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(7):4668–73. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, et al. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. Journal of Infectious Diseases. 2005;192(5):760–70. . [DOI] [PubMed] [Google Scholar]
- 10. McShan WM, Ferretti JJ, Karasawa T, Suvorov AN, Lin S, Qin B, et al. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes . Journal of Bacteriology. 2008;190:7773–85. JB.00672-08 [pii] 10.1128/JB.00672-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bessen DE, Kumar N, Hall GS, Riley DR, Luo F, Lizano S, et al. Whole-genome association study on tissue tropism phenotypes in group A streptococcus. Journal of bacteriology. 2011;193(23):6651–63. Epub 2011/09/29. 10.1128/JB.05263-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fittipaldi N, Beres SB, Olsen RJ, Kapur V, Shea PR, Watkins ME, et al. Full-Genome dissection of an epidemic of severe invasive disease caused by a hypervirulent, recently emerged clone of group A Streptococcus . The American journal of pathology. 2012;180(4):1522–34. 10.1016/j.ajpath.2011.12.037 [DOI] [PubMed] [Google Scholar]
- 13. Desiere F, McShan WM, van Sinderen D, Ferretti JJ, Brussow H. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic Streptococci: evolutionary implications for prophage-host interactions. Virology. 2001;288(2):325–41. 10.1006/viro.2001.1085 . [DOI] [PubMed] [Google Scholar]
- 14. Canchaya C, Desiere F, McShan WM, Ferretti JJ, Parkhill J, Brussow H. Genome analysis of an inducible prophage and prophage remnants integrated in the Streptococcus pyogenes strain SF370. Virology. 2002;302(2):245–58. 10.1006/viro.2002.1570 [DOI] [PubMed] [Google Scholar]
- 15. Scott J, Thompson-Mayberry P, Lahmamsi S, King CJ, McShan WM. Phage-associated mutator phenotype in group A Streptococcus . Journal of Bacteriology. 2008;190:6290–301. 10.1128/JB.01569-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scott J, Nguyen S, King CJ, Hendrickson C, McShan WM. Mutator Phenotype Prophages in the Genome Strains of Streptococcus pyogenes: Control by Growth State and by a Cryptic Prophage-Encoded Promoter. Frontiers in Microbiology. 2012;3 10.3389/fmicb.2012.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus . Mol Microbiol. 1998;29(2):527–43. . [DOI] [PubMed] [Google Scholar]
- 18. Novick RP, Christie GE, Penades JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol. 2010;8(8):541–51. Epub 2010/07/17. doi: nrmicro2393 [pii] 10.1038/nrmicro2393 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Euler CW, Juncosa B, Ryan PA, Deutsch D, McShan WM, Fischetti VA. Targeted Curing of All Lysogenic Bacteriophage from Streptococcus pyogenes using a Novel Counter-selection Technique. Submitted. 2015. [DOI] [PMC free article] [PubMed]
- 20. Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 2011;7(8):e1002190 10.1371/journal.ppat.1002190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2(1):e5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kjems E. Studies on streptococcal bacteriophages. 3. Hyaluronidase produced by the streptococcal phage-host cell system. Acta Pathologica et Microbiologica Scandinavica. 1958;44(4):429–39. 10.1111/j.1699-0463.1958.tb01094.x [DOI] [Google Scholar]
- 23. Pitcher DG, Saunders NA, Owen RJ. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Letters in Applied Microbiology. 1989;8:151–6. [Google Scholar]
- 24. Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Third ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 25. Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1948;28:491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20(1):4–17. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shaver AC, Sniegowski PD. Spontaneously arising mutL mutators in evolving Escherichia coli populations are the result of changes in repeat length. Journal of bacteriology. 2003;185(20):6076–82. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazurkiewicz P, Poelarends GJ, Driessen AJ, Konings WN. Facilitated drug influx by an energy-uncoupled secondary multidrug transporter. J Biol Chem. 2004;279(1):103–8. . [DOI] [PubMed] [Google Scholar]
- 29. Winkle SA, Rosenberg LS, Krugh TR. On the cooperative and noncooperative binding of ethidium to DNA. Nucleic Acids Res. 1982;10(24):8211–23. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 31. McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, et al. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41(14):e140 10.1093/nar/gkt444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malhotra-Kumar S, Van Heirstraeten L, Lammens C, Chapelle S, Goossens H. Emergence of high-level fluoroquinolone resistance in emm6 Streptococcus pyogenes and in vitro resistance selection with ciprofloxacin, levofloxacin and moxifloxacin. J Antimicrob Chemother. 2009;63(5):886–94. 10.1093/jac/dkp057 [DOI] [PubMed] [Google Scholar]
- 33. Pletz MW, McGee L, Van Beneden CA, Petit S, Bardsley M, Barlow M, et al. Fluoroquinolone resistance in invasive Streptococcus pyogenes isolates due to spontaneous mutation and horizontal gene transfer. Antimicrob Agents Chemother. 2006;50(3):943–8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma W, Sandri GvH, Sarkar S. Analysis of the Luria-Delbrück distribution using discrete convolution powers. J Appl Prob. 1992; 29:255–67. [Google Scholar]
- 35. Stewart FM. Fluctuation tests: how reliable are the estimates of mutation rates? Genetics. 1994;137(4):1139–46. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–32. Epub 1982/05/05. doi: 0022-2836(82)90515-0 [pii]. . [DOI] [PubMed] [Google Scholar]
- 37. Bolhuis H, Poelarends G, van Veen HW, Poolman B, Driessen AJ, Konings WN. The Lactococcal lmrP gene encodes a proton motive force-dependent drug transporter. J Biol Chem. 1995;270(44):26092–8. . [DOI] [PubMed] [Google Scholar]
- 38. Putman M, van Veen HW, Degener JE, Konings WN. The lactococcal secondary multidrug transporter LmrP confers resistance to lincosamides, macrolides, streptogramins and tetracyclines. Microbiology. 2001;147(Pt 10):2873–80. . [DOI] [PubMed] [Google Scholar]
- 39. Friedberg EC, Walker GC, Siede W, Wood RD, Schutlz RA, Ellenberger T. DNA Repair and Mutagenesis. Second ed. Washington, D.C.: ASM Press; 2006. 1118 p. [Google Scholar]
- 40. Coulondre C, Miller JH. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli . J Mol Biol. 1977;117(3):577–606. Epub 1977/12/15. doi: 0022-2836(77)90059-6 [pii]. . [DOI] [PubMed] [Google Scholar]
- 41. Grzesiuk E, Janion C. Some aspects of EMS-induced mutagenesis in Escherichia coli . Mutat Res. 1993;297(3):313–21. Epub 1993/10/01. . [DOI] [PubMed] [Google Scholar]
- 42. Caparon MG, Stephens DS, Olsen A, Scott JR. Role of M protein in adherence of group A streptococci. Infect Immun. 1991;59(5):1811–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neyfakh AA, Borsch CM, Kaatz GW. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob Agents Chemother. 1993;37(1):128–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watson ME Jr., Nielsen HV, Hultgren SJ, Caparon MG. Murine vaginal colonization model for investigating asymptomatic mucosal carriage of Streptococcus pyogenes . Infect Immun. 2013;81(5):1606–17. 10.1128/IAI.00021-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Paluscio E, Caparon MG. Streptococcus pyogenes Malate Degradation Pathway Links pH Regulation and Virulence. Infect Immun. 2015;83(3):1162–71. 10.1128/IAI.02814-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen–host interactions. Trends in Microbiology. 2003;11(5):224–32. 10.1016/s0966-842x(03)00098-2 [DOI] [PubMed] [Google Scholar]
- 47. Hause LL, McIver KS. Nucleotides critical for the interaction of the Streptococcus pyogenes Mga virulence regulator with Mga-regulated promoter sequences. J Bacteriol. 2012;194(18):4904–19. 10.1128/JB.00809-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cusumano ZT, Caparon MG. Citrulline protects Streptococcus pyogenes from Acid Stress Using the Arginine Deiminase Pathway and the F1F0 ATPase. J Bacteriol. 2015. 10.1128/JB.02517-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kietzman CC, Caparon MG. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect Immun. 2010;78(1):241–52. Epub 2009/10/21. doi: IAI.00746-09 [pii] 10.1128/IAI.00746-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carroll RK, Musser JM. From transcription to activation: how group A streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol Microbiol. 2011;81(3):588–601. 10.1111/j.1365-2958.2011.07709.x . [DOI] [PubMed] [Google Scholar]
- 51. Chaussee M, Phillips E, Ferretti J. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect Immun. 1997;65(5):p1956–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chaussee M, Ajdic D, Ferretti J. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect Immun. 1999;67(4): 1715–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lyon W, Gibson C, Caparon M. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes . EMBO J. 1998;17(21):6263–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rudan M, Schneider D, Warnecke T, Krisko A. RNA chaperones buffer deleterious mutations in E. coli . eLife. 2015;4 10.7554/eLife.04745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barksdale L, Arden SB. Persisting bacteriophage infections, lysogeny, and phage conversions. Annual Review of Microbiology. 1974;28:265–99. 10.1146/annurev.mi.28.100174.001405 . [DOI] [PubMed] [Google Scholar]
- 56. Wagner PL, Waldor MK. Bacteriophage control of bacterial virulence. Infect Immun. 2002;70(8):3985–93. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brussow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68(3):560–602. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McShan WM, Ferretti JJ. Bacteriophages and the Host Phenotype In: McGrath S, editor. Bacteriophages: genetics and molecular biology. Hethersett, Norwich, UK: Horizon Scientific Press; 2007. p. 229–50. [Google Scholar]
- 59. Kreikemeyer B, Nakata M, Koller T, Hildisch H, Kourakos V, Standar K, et al. The Streptococcus pyogenes serotype M49 Nra-Ralp3 transcriptional regulatory network and its control of virulence factor expression from the novel eno ralp3 epf sagA pathogenicity region. Infect Immun. 2007;75(12):5698–710. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marks LR, Mashburn-Warren L, Federle MJ, Hakansson AP. Streptococcus pyogenes biofilm growth in vitro and in vivo and its role in colonization, virulence, and genetic exchange. J Infect Dis. 2014;210(1):25–34. 10.1093/infdis/jiu058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mirouze N, Berge MA, Soulet AL, Mortier-Barriere I, Quentin Y, Fichant G, et al. Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci U S A. 2013;110(11):E1035–44. 10.1073/pnas.1219868110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Berge M, Moscoso M, Prudhomme M, Martin B, Claverys JP. Uptake of transforming DNA in Gram-positive bacteria: a view from Streptococcus pneumoniae . Mol Microbiol. 2002;45(2):411–21. . [DOI] [PubMed] [Google Scholar]
- 63. Nguyen SV, McShan WM. Chromosomal Islands of Streptococcus pyogenes and related streptococci: Molecular Switches for Survival and Virulence. Frontiers in Cellular and Infection Microbiology. 2014;4 10.3389/fcimb.2014.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gonzalez K, Faustoferri RC, Quivey RG Jr. Role of DNA base excision repair in the mutability and virulence of Streptococcus mutans . Mol Microbiol. 2012;85(2):361–77. 10.1111/j.1365-2958.2012.08116.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phage-like chromosomal island SpyCIM1 regulates the expression of an operon containing genes mutL, lmrP, ruvA, and tag in response to cell growth [15, 16]. The presence of an integrated SpyCIM1 results in the cells adopting a mutator phenotype with regard to DNA mismatch repair (MMR), multiple drug efflux, Holliday junction resolution, and base excision repair. In early logarithmic phase, SpyCIM1 excises from the SF370 chromosome, restoring expression of mutL and the downstream genes (insert). As the cells approach stationary phase, SpyCIM1 re-integrates into mutL, silencing gene expression. This process of SpyCIM1 excision and integration causes the cells to alternate between a wild type and mutator phenotype. In the figure, transcriptionally active MMR operon genes are black while inactive ones are gray. The identification and potential function of the SpyCIM1 genes are detailed in S1 Table). Adapted from Nguyen and Scott [63], and used by permission.
(TIF)
Strains SF370SmR and CEM1Δ4 were grown overnight at 37°C in CDM. Gentle shaking was used to simultaneously disperse each cell pellet, showing that SF370SmR had a clumping phenotype that CEM1Δ4 lacked. The cultures were streaked on blood agar to confirm that both were pure cultures of each starting strain.
(TIF)
The data are taken from Fig 5. The first gene in the MMR operon, mutS, is not shown since no differences in transcription were ever observed between SF370SmR and CEM1Δ4. Only differences in transcription ≥ ±3-fold are shown.
(TIF)
Transcriptome analysis of mRNA from SF370SmR allowed prediction of the messages encoded by SpyCIM1, including probable polycistronic ones. The genetic map of SpyCIM1 is shown flanked by the MMR operon genes. Row A below the genetic map shows the predicted mRNAs that match genes in the SF370SmR annotation. Row B shows small RNAs that were detected by RNA-Seq but which are not included in the Genbank annotation. The small RNA immediately upstream of the int mRNA encodes a small peptide with a transmembrane domain. Prediction was accomplished using the software package Rockhopper [31].
(TIF)
The protein IDs refer to the ORF numbers in the published SF370 genome [1]. Products are predicted by BLASTP homology of the encoded proteins to GenBank entries. Several additional ORFs have been identified since the original annotation was prepared.
(PDF)
A likelihood ratio test was used to calculate the ratio of SF370SmR RNA to CEM1Δ4 RNA for each gene in the SF370 Genbank annotation. A Benjamini and Hochberg correction was applied to the data using the software package GeneSifter. Ratios greater than 3 or less than -3 are reported and were used to create the plot in Fig 5.
(PDF)
The analysis was done using GeneSifter as above.
(PDF)
The analysis was done using GeneSifter as above.
(PDF)
The analysis was done using GeneSifter as above.
(PDF)
The analysis was done using GeneSifter as above.
(PDF)
The cDNA preparations used for RNA-seq analysis were analyzed by qRT-PCR for comparing the expression of nga, slo, norA, emm1, speB, and hasB in SF370SmR to CEM1Δ4 as described in the Methods.
(PDF)
Three independent cultures of SF370SmR and of CEM1Δ4 were grown at 39°C; samples were harvested for RNA isolation when the culture density (A600 nm) was 0.2 (EL) and again when the density was 0.5 (LL). An addition sample was harvested one-hour post LL (Stationary). After conversion of the RNA to cDNA, qRT-PCR was used to compare expression levels of the listed genes between SF370SmR and CEM1Δ4. Values are the average and standard deviation of the fold-difference between the two stains.
(PDF)
Data Availability Statement
Data are available in the GEO database, accession record GSE75633 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75633).