Fig 1. Key steps in the origin-dependent inverted-repeat amplification model for the generation of interstitial, inverted triplications.
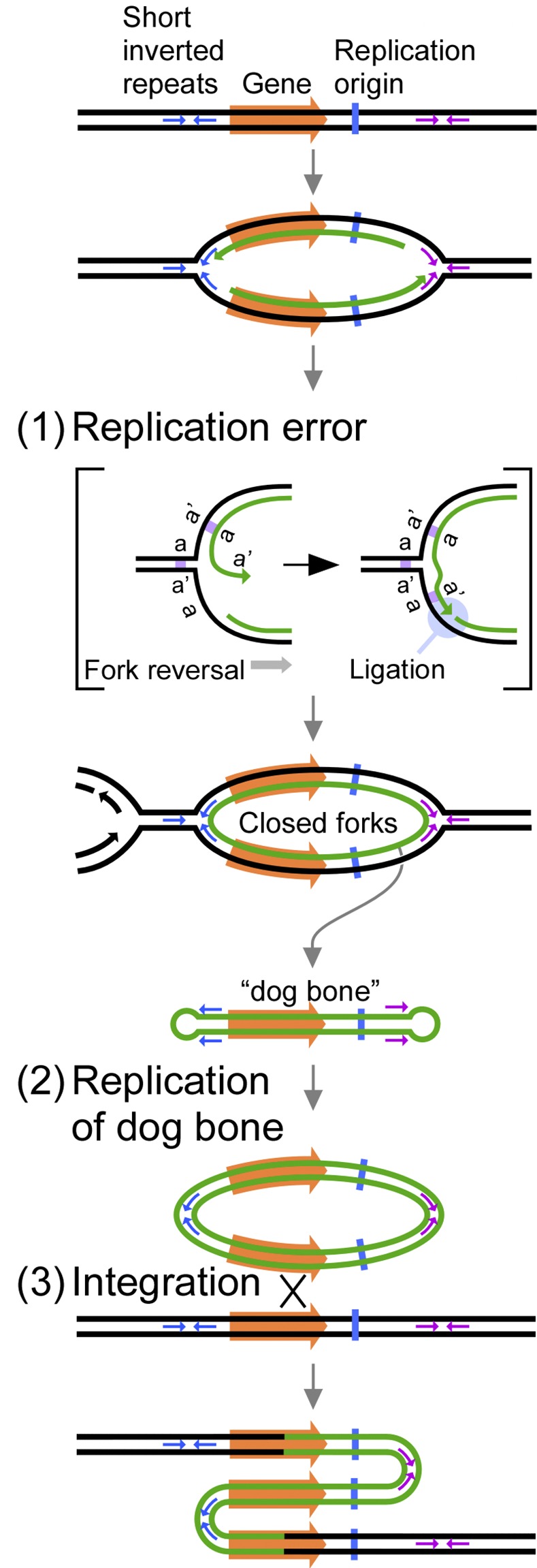
(1) Replication forks initiated at an origin of replication (vertical blue line) undergo fork reversal at small, interrupted inverted repeats (small blue/purple arrows; a and a’) leading to a replication error that covalently links nascent leading and lagging strands. These self-complementary DNA loops are expelled by adjacent replication forks to generate free linear DNA duplexes with closed loops (“dog bone” molecules) that correspond to the interrupted, inverted repeats and their intervening sequences. (2) In the next cell cycle, replication from the origin on the linear fragment generates a circular plasmid with the two copies of the amplified region in inverted orientation with the interrupted, inverted repeats at the head-to-head and tail-to-tail junctions. (3) Integration of the plasmid into the chromosomal locus generates the triplication with the central repeat in reverse orientation. The drawings are adapted from Fig 2 in [2].
