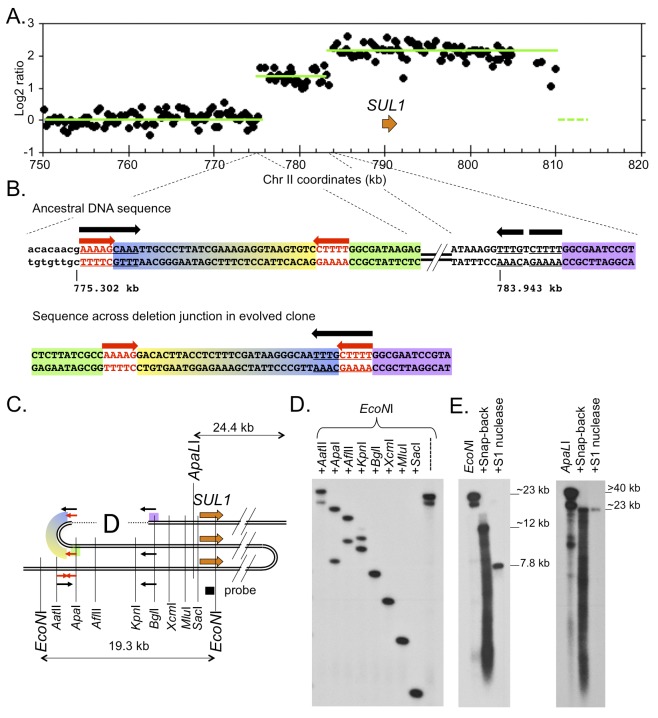
Fig 8. Secondary rearrangement at a palindromic junction in a SUL1 amplicon.
(A) ArrayCGH of yeast strain 10206-c1476. Only the last 65 kb of chromosome II are shown. (Due to the lack of unique probes near the right telomere, it is not possible to determine from aCGH the precise right junction of the amplicon.) (B) Sequences of the ancestral chromosome and the rearranged palindromic junction from the evolved clone. Sequences centromere-proximal of the inversion junction are in lower case. The short inverted repeats proposed to be the site of fork closure are indicated in red font (red arrow); sequences believed to be the sites of intramolecular, non-allelic, homologous recombination to generate the deletion of one arm of the palindrome are underlined (black arrow). Colored highlights are included to illustrate the orientation and junctions created after formation of the amplicon and the deletion of one of the palindromic arms. (C) Map illustrating the structure of the original inferred inverted triplication, highlighting the short inverted repeats (red arrows) and the sites of homology (black arrows). Colored blocks are the same as in (B). The direct orientation of the two distal black arrows creates an opportunity for intramolecular, non-allelic, homologous recombination to remove the intervening sequences. (D) Confirmation of the structure of the centromere-proximal amplicon junction by indirect end-labeling. DNA from 10206-c1476 was digested with EcoNI, distributed to 9 tubes and digested with one of the indicated enzymes. A probe (black square) adjacent to the EcoNI site detects fragments that extend from the EcoNI site toward the centromere. The map in (C) illustrates the locations of sites in the ancestral genome; these fragment sizes are detected for the most centromere proximal copy of the SUL1 locus and for fragments completely contained within the two arms of the inverted amplicon. The second bands seen for ApaI, AflII, and KpnI are consistent with sizes expected for fragments that span the deletion. (E) Snap-back assays on EcoNI and ApaLI digested genomic DNA from 10206-c1476. Both ancestral and amplicon specific fragments are detected in native DNA. Denaturation and quick cooling produces a smear of ssDNA fragments along with duplex hairpins of approximately 12 kb for the EcoNI digest and 23 kb for the ApaLI digest. There is no change in size of the ApalI fragment after S1 nuclease digestion, but only a 7.8 kb duplex portion of the EcoNI hairpin remains after S1 digestion. The change in size is consistent with the EcoNI hairpin having lost a large single-stranded loop during S1 digestion.