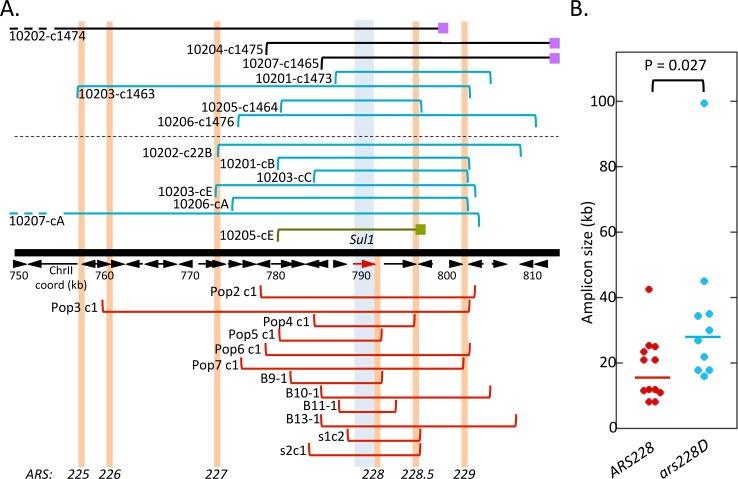
Fig 9. SUL1 amplicon structures generated in the presence and absence of ARS228.
(A) Top: Seven sulfate limited chemostats were grown for ~200 generations with a strain deleted for ARS228. A single random clone was isolated from each chemostat and characterized by ApaLI snap-back assays (S5 Fig) and aCGH (the amplification junctions are marked by brackets above the dotted line, aligned with the chromosomal map of the right arm of chromosome II from coordinates 750 kb to telomere (813 kb)). Among the single clones (S10201 to 7) we identified four clones with interstitial inverted amplification events (blue brackets) and three clones which were inconsistent with inverted amplification structures (black brackets; 10202-c1474 also suffered a telomere deletion). Additional unique clones (designated as cA-cE, below the dotted line) were analyzed from each chemostat culture. Among the seven additional clones, one clone was a complex isochromosome with deletion of one copy of CEN2 (green clone). The junction sequence (green square) was not pursued further. (A) Bottom: Wild type cultures (ARS228) selected from the sulfate-limited chemostat cultures (~200 generations) were pooled from published work from our labs and their amplification junctions are shown below the chromosome map (red brackets). Molecular characterizations of chromosome II structures in these clones are consistent with interstitial inverted amplification. Pop 2–7 clones are from [4]; B9, 10, 11, and 13 are from [18]; and s1c2 and s2c1 are from [16]. The vertical orange bars mark the positions of known ARSs. (B) Sizes of amplified regions from the ARS228 and ars228Δ strains generate medians of 16.5 and 28.9 kb, respectively, with a Wilcoxon Mann Whitney significance p-value of 0.027.