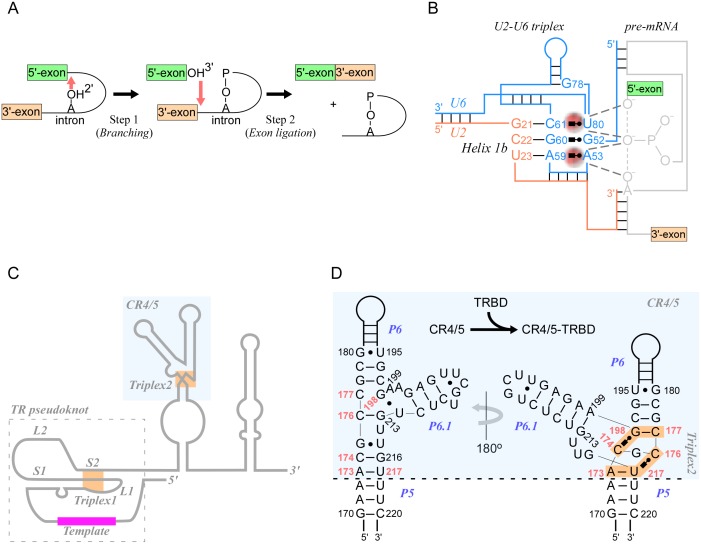
Fig 4. RNA triplexes perform critical functions in biological systems.
(A) Schematic of the two catalytic steps of splicing. In the first step (branching), the 2′-hydroxyl of an intronic adenosine (branch point) attacks the phosphodiester bond at the 5′-exon-intron boundary, releasing the 5′-exon with a free 3′-hydroxyl and a lariat structure comprising the 5′-intronic phosphate group linked to the 2′-hydroxyl of the attacking adenosine. In the second step (exon ligation), the 3′-hydroxyl of the free 5′-exon attacks the 3′-intron-exon boundary, thereby releasing the intron lariat and the fused 5′-to-3′-exons. (B) Illustration of the triplex formed by U2-U6 RNAs of yeast spliceosome. Bases from U6 (blue) and U2 (orange) create a triplex structure that coordinates two magnesium ions (red) required for both steps of catalysis on pre-mRNAs (gray). The first catalytic reaction is shown, i.e., attack of the 2′-OH of an intronic adenosine on the 5′-exon-intron junction phosphate group. (C) Outline of the TR component of telomerase (medaka), displaying the core pseudoknot region comprising two loops (L1 and L2) and two stems (S1 and S2), template (red), and two triplexes (orange), one at the pseudoknot and the other at the CR4/5 domain (blue shading). (D) Close-up of the CR4/5 domain showing base-pair interactions in the absence (left) and in the presence (right) of the TR-binding domain (TRBD). TRBD binding reorganizes critical bases (red) at the junction between P5, P6, and P6.1 and form a mini-triplex, causing P6.1 to rotate by approximately 180° (blue shading).