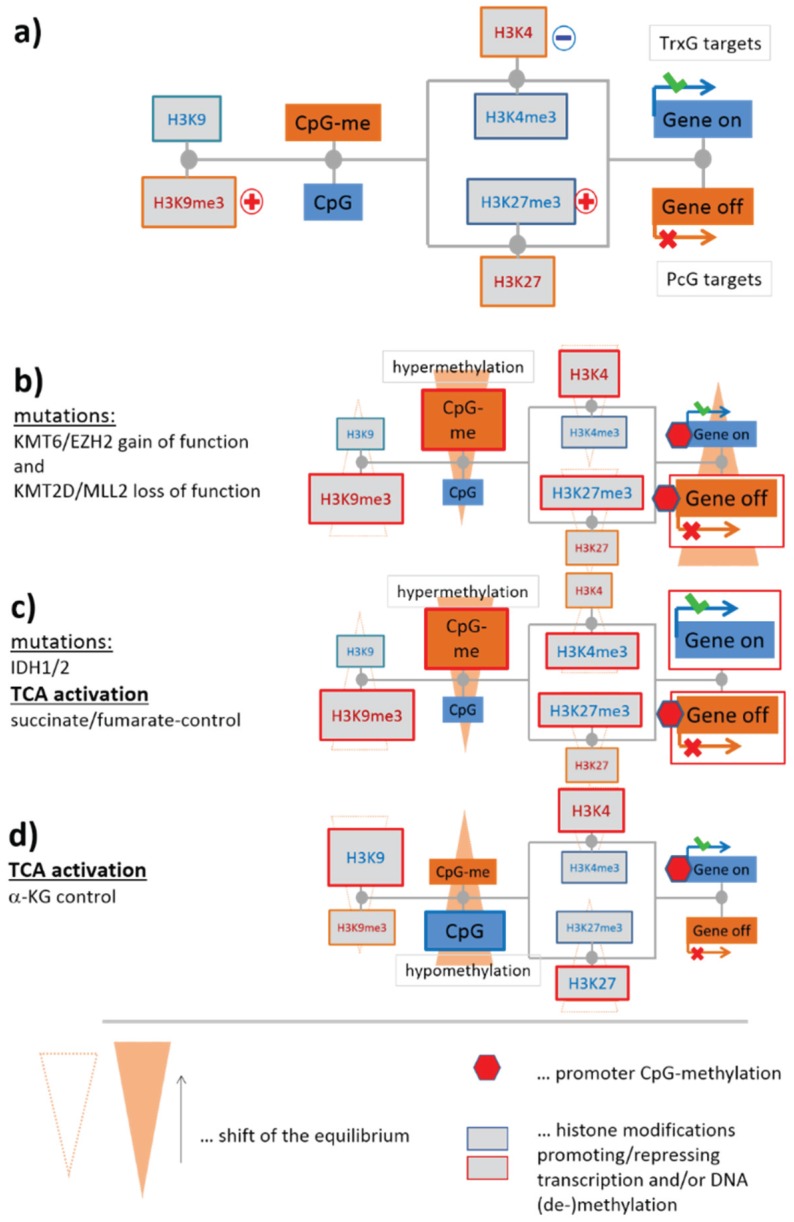
Figure 4.
Effect of selected mutations of epigenetic modifiers and of tricarboxylic acid (TCA) metabolism on CpG methylation and gene expression: (a) Simplified sketch of the scheme shown in Figure 3. The plusses and the minus indicate gain and loss of function mutations, respectively; (b) The scheme suggests that mutations of EZH2 and MLL2 result in DNA-hypermethylation and de-activation of transcription. Alterations are shown by enlarged boxes and red frames in the simplified scheme; (c) IDH1 mutations will inhibit a series of KDMs via 2-HG produced by mutated IDH1. It results in hypermethylation and alterations of gene expression in both directions. The same trend is expected for increased TCA-activities with increased levels of fumarate and succinate. Both metabolites inhibit the same modifiers as 2HG; (d) TCA activation with increased amounts of α-KG will have the opposite effect as in (c): it will demethylate DNA and downregulate expression. Note that global changes of methylation (as indicated by “CpG-me”) are paralleled by local ones due to the hypothesized alterations of gene expressions: red marks indicate promoter methylations in (b–d) recruited by polycomb group (PcG)-repressed and/or trithorax group (TrxG)-deactivated genes.