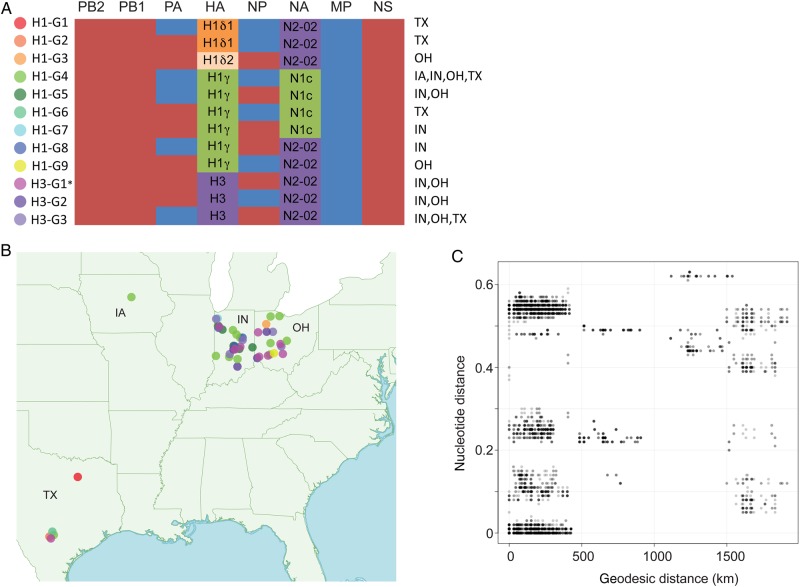
Figure 3.
Twelve influenza A virus genotypes identified in exhibition swine in 2013. A, The evolutionary origins of each of the 6 internal gene segments of the influenza A virus genome (polymerase basic 2 [PB2], PB1, polymerase acidic [PA], nucleoprotein [NP], matrix protein [MP], and nonstructural protein [NS]) are classified as triple-reassortant internal genes (red) or 2009 pandemic influenza A(H1N1) (blue). The hemagglutinin (HA) segments are classified as H1δ1 (H1d1; orange), H1δ2 (H1d2, shaded beige), H1γ (H1 g, shaded green), or H3 (shaded purple). The neuraminidase (NA) segments are classified as classical N1 (N1C; green) or N2-2002 (N2-02; purple). The A(H1N1)/A(H1N2) genotypes are numbered H1-G1 to H1-G9. The A(H3N2) genotypes are numbered H3-G1 to H3-G3, consistent with genotype nomenclature used previously [10]. The asterisk indicates that genotype H3-G1 viruses represents the genotype responsible for variant A(H3N2) cases in humans that occurred during 2011–2012. The US states in which each genotype was identified in exhibition swine in 2013 are specified. B, The global positioning system–based location where viruses of each genotype were identified in exhibition swine in 2013 is provided, with the shading of solid circles corresponding to those provided in panel A. C, Plot of geodesic distances (km) vs genetic distances (nucleotide substitutions per site) between influenza viruses collected from exhibition swine in 2013.