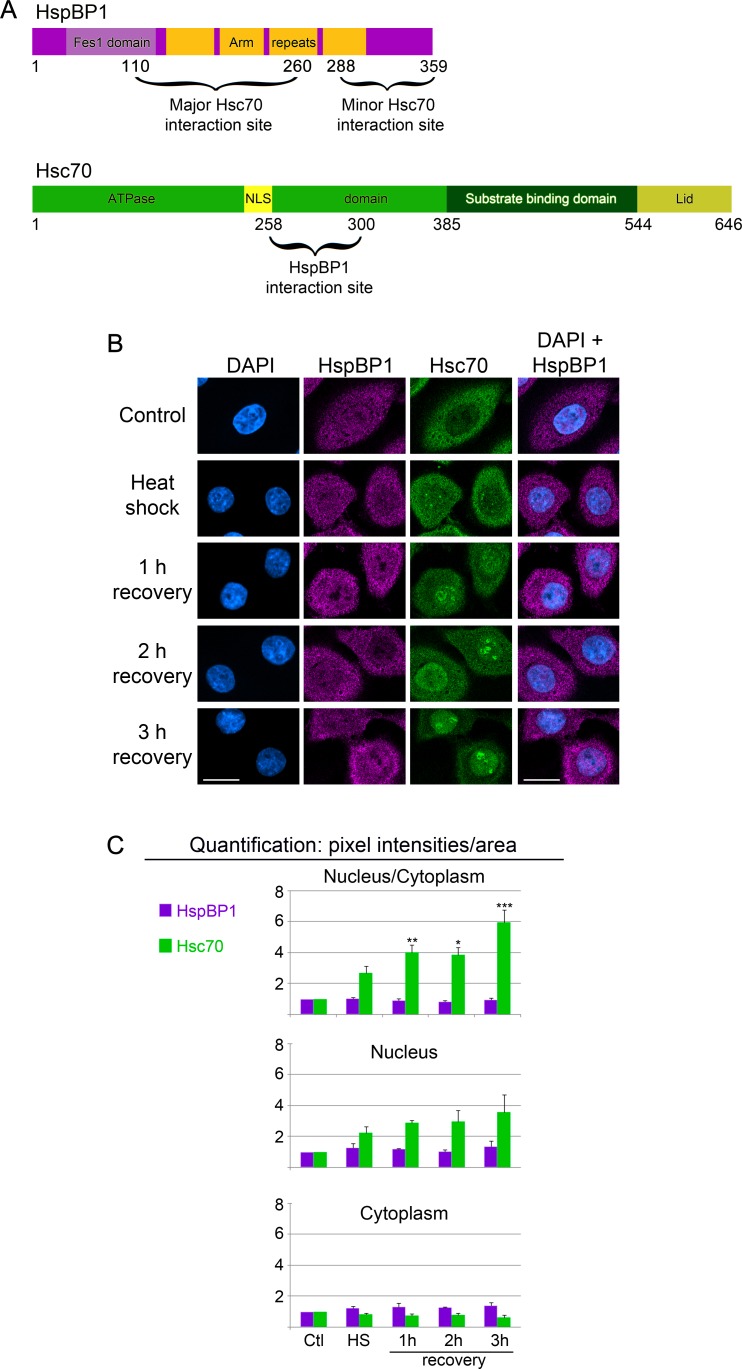
Figure 1. Subcellular distribution of the co-chaperone HspBP1 and its binding partner hsc70 under control and stress conditions.
(A) HspBP1 and hsc70 organization. Domains are depicted for HspBP1 and hsc70, numbers denote amino acid residues. The regions involved in the co-chaperone/chaperone interactions are marked; they are based on the crystal structure (Shomura et al., 2005). NLS denotes the nucleolar localization sequence (Banski et al., 2010). Hsc70 and HspBP1can be modified posttranslationally; some of these modifications occur in the chaperone/co-chaperone interaction sites (Table 1, (Hornbeck et al., 2015)). (B) HeLa cells were grown at 37 °C or exposed to heat shock, followed by stress recovery. Representative confocal images for the immunolocalizations of HspBP1 and hsc70 are shown. Nuclei were detected with DAPI; an overlay of DAPI and HspBP1 images demarcates the nuclear compartments. Size bars are 20 µm. (C) Pixel intensities were quantified in the nucleus or cytoplasm, and the nucleocytoplasmic ratio was calculated. Results are shown as means + SEM for three independent experiments. Significant differences were identified by One-Way ANOVA, using unstressed control cells as the reference; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.