Abstract
Recent findings suggest diverse and potentially multiple roles of SUMO in testicular function and spermatogenesis. However, SUMO targets remain uncharacterized in the testis due to the complex multicellular nature of testicular tissue, the inability to maintain and manipulate spermatogenesis in vitro, and the technical challenges involved in identifying low-abundance endogenous SUMO targets. In this study, we performed cell-specific identification of sumoylated proteins using concentrated cell lysates prepared with de-sumoylation inhibitors from freshly purified spermatocytes and spermatids. One-hundred and twenty proteins were uniquely identified in the spermatocyte and/or spermatid fractions. The identified proteins are involved in the regulation of transcription, stress response, microRNA biogenesis, regulation of major enzymatic pathways, nuclear-cytoplasmic transport, cell cycle control, acrosome biogenesis, and other processes. Several proteins with important roles during spermatogenesis were chosen for further characterization by co-immunoprecipitation, co-localization and in-vitro sumoylation studies. GPS-SUMO software was used to identify consensus and non-consensus sumoylation sites within the amino acid sequences of the proteins. The analyses confirmed the cell-specific sumoylation and/or SUMO interaction of several novel, previously uncharacterized SUMO targets such as CDK1, RNAP II, CDC5, MILI, DDX4, TDP-43 and STK31. Furthermore, several proteins that were previously identified as SUMO targets in somatic cells (e.g., KAP1, MDC1) were identified as SUMO targets in germ cells. Many of these proteins have a unique role in spermatogenesis and during meiotic progression. This research opens a novel avenue for further studies of SUMO at the level of individual targets.
Introduction
Spermatogenesis includes the proliferation of spermatogonia through mitosis, the production of round spermatids from spermatocytes by meiosis, and the post-meiotic maturation of spermatids, termed spermiogenesis. Abnormalities during any of these processes can result in the production of malfunctioning sperm, which may lead to infertility, spontaneous abortion, or birth defects. These facts emphasize the need for a better understanding of spermatogenesis and its regulation, particularly through the characterization of the molecules that have not been well studied in the testis but that regulate important pathways in other tissues.
Various protein post-translational modifications (PTMs) regulate spermatogenesis, one of which is the attachment of small ubiquitin-like modifiers (SUMOs) in a process termed sumoylation. SUMOs are structurally similar to ubiquitin. However, their amino acid sequences differ greatly, with only ~18% similarity (Bayer et al. 1998). Four SUMO paralogs have been identified in mammals: SUMO1, 2, 3 and 4. SUMO1 (also named sentrin) shares approximately 50% homology with SUMO 2 and SUMO3, which are usually referred to as SUMO2/3 given that they are 95% identical. During sumoylation, an isopeptide bond forms between a SUMO and the lysine residue of its substrate. This process requires a SUMO-activating enzyme (E1), a SUMO-conjugating enzyme (E2) and a SUMO ligase (E3) (Yeh et al. 2000, Wang & Dasso 2009, Yeh 2009, Wilkinson & Henley 2010). Sumoylation often occurs on a target lysine residue within the consensus sequence: ψ-K-X-D/E, where ψ is a hydrophobic amino acid and X can be any amino acid (Rodriguez et al. 2001). However, not all consensus sequences are sumoylated, and sumoylation often occurs outside of the consensus sequence (Blomster et al. 2010). Notably, SUMO2/3 but not SUMO1 contain the consensus sequence, and mixed SUMO chains with a terminal SUMO1 have been reported (Rodriguez et al. 2001). Sumoylation is a dynamic process that can be reversed through the activity of sentrin-specific proteases (SENPs) by the cleavage of the isopeptide bond between the SUMO moiety and the substrate (Mukhopadhyay & Dasso 2007, Yeh 2009, Hannoun et al. 2010, Wilkinson & Henley 2010). A diverse set of SUMO target proteins has been identified in somatic cells, including factors that regulate transcription, replication, DNA repair, RNA metabolism, translation, and cellular transport. In addition to the numerous targets of sumoylation that have been identified, there is a growing list of proteins that interact with SUMO non-covalently (Song et al. 2004, Chupreta et al. 2005, Song et al. 2005, Lin et al. 2006, Kerscher 2007).
Several developmental processes, including spermatogenesis, have been studied in SUMO1-knockout mice (Zhang et al. 2008). In contrast to a previously published study that reported abnormal development of the palate in SUMO1 knockouts (Alkuraya et al. 2006), Zhang et al. found no abnormalities in mouse development, suggesting that SUMO2 and SUMO3 compensated for the functions of SUMO1. Although these results require further evaluation, they suggest that the SUMO-conjugating machinery, and not an individual SUMO isoform, should be the target of future experiments aiming to inhibit sumoylation. Unfortunately, knockout mice for UBC9, a SUMO-conjugating enzyme, show early embryonic lethality and severe disruptions in mitosis, a finding that supports the indispensable role of sumoylation in mitotic progression (Nacerddine et al. 2005). We and other groups have studied SUMO1 and SUMO2/3 in mouse and human spermatogenesis using advanced cell-imaging techniques and immunodetection analyses. SUMO localized to spermatogonia, the sex chromosomes and the centromeric heterochromatin of spermatocytes, the chromocenters of round spermatids, the centrosome area of elongating spermatids, and the nuclei of testicular somatic cells. These findings are consistent with the diverse and potentially multiple roles of SUMO in testicular function and spermatogenesis, such as spermatogonia proliferation, meiotic sex chromosome inactivation, centromeric heterochromatin organization, and reshaping the spermatid nucleus. (Rogers et al. 2004, Vigodner & Morris 2005, Vigodner et al. 2006, Brown et al. 2008, Metzler-Guillemain et al. 2008, Vigodner 2009). In agreement with these results, one study (La Salle et al. 2008) showed the dynamic nature of the gene expression levels related to sumoylation during spermatogenesis. Recent studies from our group also revealed changes in global sumoylation following the induction of various stresses in germ cells and sperm (Shrivastava et al. 2010, Shrivastava et al. 2014). Although these studies provided important initial information about the possible roles of sumoylation in spermatogenesis, little progress has been made in understanding how SUMO regulates the suggested functions. As has been shown in somatic cells, the identification of targets for sumoylation is a critical step toward understanding its cellular functions (Andersen et al. 2009, Golebiowski et al. 2009, Sarge & Park-Sarge 2009, Tatham et al. 2011). TOP2A and synaptonemal complex proteins (SYCP1 and SYCP2) have been co-immunoprecipitated with SUMO from testicular lysates, as shown by our group (Shrivastava et al. 2010) and others (Brown et al. 2008), respectively. Hundreds of SUMO targets have been identified in somatic cells. However, with the exception of TOP2A and SYCPs, SUMO targets remain uncharacterized in the testis. This knowledge gap is due to the complex, multicellular nature of testicular tissue, the inability to maintain and manipulate spermatogenesis in vitro, and the technical challenges involved in identifying low-abundance endogenous SUMO targets. To overcome some of these difficulties, we recently optimized the identification of sumoylated proteins using concentrated cell lysates, isopeptidase inhibitors to prevent de-sumoylation, and a large amount of anti-SUMO antibody crosslinked to agarose beads (Xiao et al. 2014). Using this approach, we recently identified a sumoylome of human sperm (Vigodner et al. 2013). Several additional sperm targets were identified by another group (Marchiani et al. 2014)). In this study, numerous sumoylated proteins were uniquely identified in the spermatocyte and/or spermatid fractions using lysates prepared from purified spermatogenic cells. Several proteins with important roles during spermatogenesis were further characterized by co-immunoprecipitation, co-localization and in-vitro sumoylation studies.
Materials and methods
Mice, cell lines, reagents and antibodies
C57BL/6NCrl mice were purchased from Charles River (Kingston, NY). The Animal Committee of Albert Einstein College of Medicine, Yeshiva University approved all animal protocols. The mouse Sertoli cell line 15P-1 (ATCC®, CRL-2618) and HEK 293 (ATCC® CRL-1573™) cells were purchased from ATCC (Manassas, VA) and grown in DMEM media with 5% fetal bovine serum (FBS, Life Technologies, 16140-071), 5% bovine growth serum (Fisher Scientific, SH30541.03), 1% penicillin/streptomycin (Life Technologies, 15140-122) and 0.5% Fungizone (Life Technologies, 15290-018) at 32°C with 5% CO2. The primary human Sertoli cell line (Lonza, MM-HSE-2305) was purchased from Lonza Group Ltd. (Walkersville, MD, USA) and cultured in the Sertoli Cell Growth Medium (SeGM) bullet kit (Lonza, 00191053) at 37°C with 5% CO2.
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise noted. All reagents for Western blotting and immunofluorescence were purchased from Life Technologies unless otherwise noted. The detergent-removal spin columns (87778), the screw-cap spin columns (69705), and the BCA protein assay kit (23227) were purchased from Thermo Scientific (Rockford, IL, USA). The whole-cell extraction kit (2910) was purchased from Millipore (Temecula, CA, USA). Information regarding the source, vendor and dilution of the antibodies used in this study is summarized in the Supplementary Table 1.
Germ cell separation by velocity sedimentation (STA-PUT)
Mouse testicular germ cells were separated using a STA-PUT sedimentation velocity cell separator (ProScience Inc., Scarborough, Ontario, Canada) and a published procedure (La Salle et al. 2009). The details of the procedure are summarized in the Supplementary materials.
Flow cytometry analysis
Chosen STAPUT fractions were centrifuged at 500 g for 7 min at 4°C followed by aspiration of all but 1 ml of supernatant. The pellets were then resuspended with a brief, gentle vortex. From each resulting cell suspension, 150 μl was individually mixed with propidium iodide (PI) staining solution (PBS with 1% (v/v) RNase A, 10 μg/ml PI, and 1% (v/v) Igepal CA-630) and incubated at 37°C for 15 min in the dark. After incubation, the samples were filtered through a nylon mesh and subjected to flow cytometric analysis of PI-stained DNA fractions (Supplementary Figure 1A). The flow cytometry and the subsequent analyses were processed by CytoSoft software (Millipore, Billerica, MA, USA). Fractions containing spermatocytes (tetraploid cells) and spermatids (haploid cells) with a purity above 80% and 90%, respectively, were pooled together for cell slides, protein extraction, or spermatocyte culture (Supplementary materials, Fig.1).
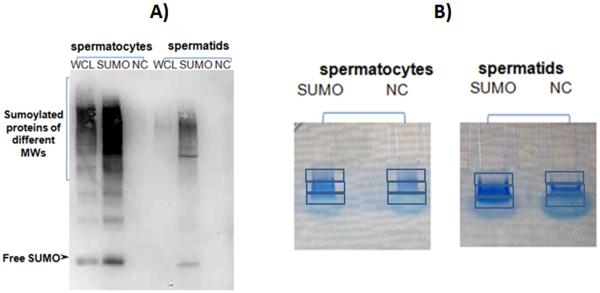
Figure 1.
(A) Western blotting confirmed the successful enrichment of the IP fraction for sumoylated proteins in spermatocytes and spermatids when compared to the negative controls. Whole-cell lysate (WCL), negative control (NC) and immunoprecipitated fraction (SUMO) are shown.
(B) The precipitated proteins and their corresponding negative controls were briefly run on gels, and the gels were subsequently fixed and stained. The stained regions were cut into three gel bands, digested and analyzed by LC-MS/MS for mass spectrometry.
Immunofluorescence (IF)
Pooled fractions were centrifuged at 500 g for 7 min and resuspended in 5 ml of Krebs Ringer Buffer (KRB) media (120 mM NaCl, 4.8 mM KCl, 25.2 mM NaHCO3, 1.2 mM KH2PO4, 1.2 mM MgSO4·7H2O, 1.3 mM CaCl2, 1 × Pen/Strep/Glu (Life technologies), 1 × essential amino acid, 1 × non-essential amino acid (Lonza, Walkersville, MD, USA), and 11.1 mM dextrose). Twenty-five microliters of the cell suspension was smeared onto a poly-L-lysine microscope slide (Polysciences Inc., 22247, Warrington, PA, USA), and the rest was subjected to protein extraction. The slides were completely air dried, and the cells were fixed with 1% paraformaldehyde (PFA) in PBS (Polysciences, 18814) for 10 min. The cells were then washed twice with PBS before being permeabilized with 0.3% Igepal CA-630 for 10 min. The cells were then blocked with Image-iT® FX signal enhancer for 30 min and then rinsed with PBS. Subsequently, the cells were incubated with the appropriate antibodies in PBS containing 1% BSA for 2 h (Supplementary Table 1). Following a PBS wash, the cells were incubated with fluorophore-conjugated secondary antibodies diluted 1:150 in PBS containing 1% BSA for 1 h. The cells were then washed, and the nuclei were stained with 4 μg/ml of 4,6-diamino-2-phenylindole (DAPI) for 5 min. After one wash, the slides were mounted with ProLong Gold antifade reagent. The images were collected with a Nikon inverted fluorescence microscope and 60 × and 100 × objective lenses with DAPI, fluorescein isothiocyanate and CY-5 filter sets. At least two slides with at least 50 cells on each slide were analyzed. The images are representative of the pattern obtained for the entire slide analyzed.
Protein extraction
Both denaturing and non-denaturing lysis buffer was used for cell lysis. Many proteins are covalently and/or non-covalently modified by SUMO. It has therefore been suggested that the use of a denaturating lysis buffer containing a high percentage of SDS would have the benefits of immediately denaturing isopeptidases and eliminating non-covalent interactions with SUMO, thereby leaving only covalent SUMO binding in place (Sarge & Park-Sarge 2009, Tatham et al. 2009, Barysch et al. 2014). The high percentage of SDS, however, would prevent sumoylated proteins from binding to anti-SUMO antibodies during the IP procedure. As a result, SDS is usually either significantly diluted (e.g., 1:10) or removed from the lysis buffer by other means (Sarge & Park-Sarge 2009, Tatham et al. 2009, Barysch et al. 2014). However, these manipulations can cause certain proteins to re-nature and non-covalent interactions to reform. In our studies of several SUMO targets, no significant difference was found between the results of a co-IP performed using denaturing and non-denaturing lysis buffers (Supplementary Fig. 2). In both cases, the conclusions concerning possible protein sumoylation were based on the presence of high-molecular weight protein conjugate/s (one or numerous) detected with both anti-SUMO and anti-target protein antibodies above the band corresponding to the non-modified form of the protein. The presence of a band corresponding to the molecular weight of the non-modified protein would suggest a non-covalent interaction.
To produce denatured lysates, cell pellets were re-suspended in modified 2 × Laemmli buffer (150 mM Tris·HCl pH 7.2, 4% sodium dodecyl sulfate (SDS), 20% glycerol and 20 mM N-ethylmaleimide (NEM, an isopeptidase inhibitor that blocks the activities of SENPs (Suzuki et al. 1999)) with 5 × 106 cells for every 300 μl of buffer. The solution was sonicated until the sample become liquid and then boiled at 100°C for 10 min. The lysates were then collected after centrifugation at high speed at room temperature for 15 min. The denaturing lysate was subjected to SDS removal using detergent-removal spin columns (Thermo Scientific) in accordance with the manufacturer's instructions. In brief, the SDS-containing lysate was slowly dropped onto the top of the compact resin of detergent-removal spin columns that had been washed with PBS and equilibrated and then incubated for two minutes at room temperature. The columns were then centrifuged at 1,000 g for two minutes at room temperature, and the flow-through fractions were collected and pooled together as the SDS-removed lysate.
To produce the non-denatured lysate, 5 × 106 cells were re-suspended in 300 μl of whole cell extraction buffer (produced from the Millipore kit) supplemented with NEM at a final concentration of 20 mM. This cell suspension was then drawn through a 27-gauge needle five times and incubated on ice for 15 min. The lysate was collected after a centrifugation at high speed at 4°C for 20 min. Lysates from 5–6 or 2–3 STA-PUT separations (for spermatocytes and spermatids, respectively) were collected to produce sufficient amounts of proteins for IP (500–1,300 μg of protein).
Immunoprecipitation (IP)
In all of the experiments, the lysates were pre-cleared by mixing them with control agarose resin (from Thermo Scientific, 26150) at 4°C for 1 h on an orbital rotator. Each 600 μl of the pre-cleared lysate (0.5–1.3 mg of protein) was mixed with 120 μl of SUMO1-agarose conjugate in a screw-cap spin column (Thermo Scientific) at 4°C. This was performed overnight on an orbital rotator and followed by centrifugation at 4000 rpm (Eppendorf, 5415C) at 4°C for 1 min. The amount of antibody was determined in preliminary experiments using an increasing antibody concentrations and analyzing its ability to precipitate the maximal amount of sumoylated proteins from lysates containing 1 mg of protein (not shown). The retained sumoylated proteins were washed twice with whole-cell extraction buffer supplemented with 20 mM NEM and then eluted with 50 ml acidic elution buffer (Thermo Scientific, 21004). Usage of the antibody-agarose conjugates prevented the heavy and light immunoglobulin chains from appearing in the eluted fraction, facilitating the subsequent mass spectrometry analysis.
For IP using other antibodies, the lysates were pre-cleared using protein A/G agarose beads and incubated with the antibodies overnight (Supplementary Table 1) Mouse or Rabbit IgG was used as a corresponding negative control. The lysates were washed as described above and incubated with protein A/G agarose beads overnight. This step was followed by additional washes and elution as described above.
Gel electrophoresis and Western Blotting
Gel electrophoresis was performed using NuPAGE 4 – 12% gradient Bis-Tris polyacrylamide gels and MOPS running buffer as previously described (Vigodner et al. 2013). The membrane (Novex nitrocellulose membrane, 0.45 μm pore size, Life Technologies) was first blocked with 2% membrane blocking agent (GE Healthcare UK Limited, RPN2125V, Little Chalfont, Buckinghamshire, UK) in PBS + 0.02% (v/v) Tween 20 (PBS-T) for 1 h at room temperature. The membrane was then incubated with primary antibodies in PBS containing 2% BSA and 0.1% sodium azide for either 2 h at room temperature or overnight at 4°C. Following three washes with PBS-T, the membrane was further incubated with secondary antibodies that were diluted to 1:5000 in PBS-T for 1 h at room temperature. The secondary antibodies used in this study included the following: ECL™ anti-rabbit IgG HRP linked (GE Healthcare UK Limited, NA934V), goat anti-mouse IgG (H + L) HRP (Millipore, AP308P), and goat anti-rat IgG-HRP (Santa Cruz, sc-2032). The image collection and quantitative analyses were performed using the Universal Hood II and Quantity One Software (Bio-Rad Laboratories, Hercules, CA). Depending on the quality of the images after protein separation followed by a Western blot with a particular antibody, the representative images for the SUMO IP results are presented either for denaturing (CDK1, STK31, and the largest subunit of RNAP II) or non-denaturing (MDC1, KAP1, MILI, DDX4, CDC5, TDP-43) protein lysates.
Gel fixation and staining
The samples to be subjected to mass spectrometry analysis were run on NuPAGE 4 – 12% gradient Bis-Tris polyacrylamide gels at 150 V for 5 min. The gels were fixed using a solution containing 50% (v/v) methanol and 7% (v/v) acetic acid at room temperature for 20 min and washed with distilled water 3 times for 5 min each. The fixed gels were then stained by incubation in GelCode® Blue Stain Reagent (Thermo Scientific, 24590) at room temperature for 1 hour. This step was followed by washing with distilled water overnight to remove excess staining solution. The stained regions were cut into 3 gel bands, digested and analyzed by LC-MS/MS for mass spectrometry.
Mass spectrometry analysis
Mass spectrometry (MS) was performed with the assistance of the Laboratory for Macromolecular Analysis and Proteomics at the Albert Einstein College of Medicine of Yeshiva University and is described below.
In-gel trypsin digestion and nanospray LC-MS/MS was performed as described in (Wang et al. 2014). Briefly, Coomassie-stained cut protein gel bands were first reduced with TCEP, alkylated with iodoacetamide and digested with trypsin. Nanospray LC-MS/MS was performed using a Linear Ion Trap (LTQ) mass spectrometer (Thermo Scientific) with the RSLC chromatography system (Thermo Scientific). The ten most intense ions determined from an initial survey scan (300–1600 m/z) were selected for fragmentation (MS/MS). The raw data files were converted to mgf text files (Mascot generic file) with Proteome Discoverer 1.2 and then merged and searched against the human or mouse NCBI database (April, 2014) using the in-house Mascot Protein Search engine (Matrix Science) with an automatic decoy database search. The following search parameters were used: trypsin, 2 missed cleavages; fixed modification of carbamidomethylation (Cys); variable modifications of deamidation (Asn and Gln), pyro-glu (Glu and Gln) and oxidation (Met); monoisotopic masses; peptide mass tolerance of 2 Da; and product ion mass tolerance of 0.6 Da. The Mascot-identified proteins were further validated with Scaffold (version 4, Proteome Software) using 99% and 95% protein and peptide probability, respectively, and a minimum of 2 peptides. The peptide and protein FDR (false discovery rates) were adjusted to 1% or less.
In vitro sumoylation assay
In vitro sumoylation assays were performed with the SUMOylation kit from Active Motif, Inc. (40120, Carlsbad, CA), following the manufacturer's protocol. The mouse GST-CDK1 recombinant protein was purchased from Sino Biological Inc. (Beijing, China).
Bioinformatics analysis
All of the putative sumoylation sites and SUMO-interactive motifs (SIMs) of the proteins were identified with SUMOsp 2.0 (The CUCKOO Workgroup, USTC). The identified proteins were divided into functional groups based on a literature search.
Results
I. Separation of spermatocytes and spermatids
The spermatocytes and spermatids were separated with a STA-PUT procedure that utilizes differential sedimentation velocity at the unit gravity of different cell types (Bellve et al. 1977, La Salle et al. 2009). The contents of the fractions collected from the separation were examined using microscopy and flow cytometry to identify tetraploid and haploid cells. The details of the procedure and the analysis of fraction purity are described in Supplementary Fig. 1 A and B.
II. Identification of sumoylated targets
For the identification of SUMO targets, proteins were extracted from isolated fractions using denaturing buffer followed by SDS removal and immunoprecipitated with anti-SUMO1-agarose conjugates as described in the Materials and Methods. Agarose resin without cross-linkage to antibodies was used as a negative control. Western blotting confirmed the successful enrichment of sumoylated proteins in the IP fraction compared with the negative controls (Fig. 1A). The precipitated proteins and their corresponding negative controls were then briefly run on gels, and the gels were subsequently fixed and stained (Fig. 1B). The stained regions were cut into three gel bands, digested and analyzed by LC-MS/MS for mass spectrometry. In addition to the enrichment of specific proteins, some non-specific background was observed in the negative controls. After protein digestion, mass spectrometry analyses revealed approximately 120 proteins uniquely in the antibody fractions but none in any of the negative controls (Table 1). These identified proteins were subdivided into eight groups according to their previously published functions (Matunis et al. 1996, Moroianu 1998, Stopka et al. 2000, Goldberg et al. 2003, Myojin et al. 2004, Cramer 2006, Stark & Taylor 2006, Bao et al. 2012, Vourekas et al. 2012, Wang et al. 2012, Lasko 2013) (Fig. 2). The largest group, with 33% –34% of the spermatocyte and spermatid SUMO targets, included proteins involved in transcription, RNA interaction and stability, and splicing. This group included numerous ribosomal and heterogeneous nuclear ribonucleoproteins, important splicing factors, and several novel SUMO targets with important roles in germ cells, such as the largest subunit of RNA polymerase II (RNAP II), DEAD (Asp-Glu-Ala-Asp) box polypeptide DDX4 and DDX42, PIWI-like protein 2 (MILI/PIWIL2), TAR DNA-binding protein -43 (TDP-43), and paraspeckles component 1 (Table 1, Fig. 2).
Table 1.
Identification of sumoylated proteins unique to the spermatocyte or spermatid IP samples via tandem mass spectrometry. Protein ID, molecular weight, and the number of unique peptides identified for each protein in spermatocytes and/or spermatids is indicated. Proteins of specific interest in the field of spermatogenesis are indicated in bold.
| Number of unique peptides | ||||
| Stress-related, heat shock proteins | Accession number * | MW | spermatocytes | spermatids |
| heat shock 70 kDa protein 4 | gi|112293266 (+3) | 94 kDa | 18 | 11 |
| inducible heat shock protein 70 | gi|118490060 (+4) | 70 kDa | 7 | |
| stress-70 protein, mitochondria – | gi|162461907 (+5) | 73 kDa | 5 | 4 |
| heat shock 70 kDa protein 1-like | gi|124339838 (+3) | 71 kDa | 2 | |
| heat shock protein 105 kDa | gi|114145505 (+5) | 96 kDa | 3 | |
| glutathione S-transferase P 1 | gi|10092608 (+5) | 24 kDa | 2 | |
| 1-Cys peroxiredoxin protein 2 | gi|3789944 (+3) | 25 kDa | 2 | |
| dnaJ homolog subfamily B member 1 (Heat shock 40 kDa protein 1) | gi|9055242 (+2) | 38 kDa | 2 | |
| protein DJ-1 | gi|55741460 (+1) | 20 kDa | 2 | |
| Ephx1 protein | gi|34784388 (+4) | 51 kDa | 2 | |
| DNA breaks, chromatin remodeling | spermatocytes | spermatids | ||
| mediator of DNA damage checkpoint protein 1 (MDC1) | gi|132626693 (+5) | 185 kDa | 8 | |
| SWI/SNF related regulator of chromatin SMARCA5 | gi|148678936 (+1) | 116 kDa | 5 | |
| SWI/SNF related regulator of chromatin SMARCA4 | gi|148693261 (+8) | 185 kDa | 2 | |
| matrin-3 | gi|25141233 (+4) | 95 kDa | 3 | 3 |
| ruvB-like | gi|9790083 (+1) | 50 kDa | 3 | |
| ruvB-like 2 | gi|6755382 (+1) | 51 kDa | 2 | |
| AT rich interactive domain 2 (ARID, RFX-like) | gi|262231796 | 196 kDa | 3 | |
| damage specific DNA binding protein 1 | gi|148709424 (+5) | 108 kDa | 3 | |
| poly [ADP-ribose] polymerase 1 | gi|20806109 (+4) | 113 kDa | 2 | |
| SUMO/ubiquitin pathway | spermatocytes | spermatids | ||
| tripartite motif protein 28 (TRIM28, KAP1) SUMO ligase | gi|148706135 (+3) | 89 kDa | 7 | 2 |
| Ubqln1 protein, partial | gi|16307349 (+8) | 47 kDa | 3 | |
| ubiquitin-associated protein 2-like isoform 3 | gi|260166704 (+9) | 117 kDa | ||
| ubiquitin-conjugating enzyme E2 N | gi|309262615 (+3) | 23 kDa | 2 | |
| ubiquitin-activating enzyme E1-like 2, isoform CRA_a | gi|148706006 (+4) | 119 kDa | 2 | |
| ubiquitin carboxyl-terminal hydrolase isozyme L3 | gi|139948802 (+1) | 26 kDa | 2 | |
| ubiq—uitin carboxy-terminal hydrolase LI | gi|148705826 (+2) | 26 kDa | 2 | |
| proteasome subunit alpha type-4 | gi|6755196 (+2) | 29 kDa | 2 | |
| proteasome subunit beta type-3 | gi|6755202 | 23 kDa | 2 | |
| 26S protease regulatory subunit 4 | gi|6679501 | 49 kDa | 2 | |
| 26S proteasome non-ATPase regulatory subunit 1 (Psmd1) | gi|116283726 (+5) | 93 kDa | 2 | |
| 26S proteasome non-ATPase regulatory subunit 2 (Psmd2) | gi|27692965 (+8) | 67 kDa | 2 | |
| 26S proteasome non-ATPase regulatory subunit 6 (Psmc6) | gi|28175479 (+3) | 44 kDa | 4 | |
| Cell cycle regulators | spermatocytes | spermatids | ||
| cyclin-dependent kinase 1 | gi|31542366 (+3) | 34 kDa | 4 | 3 |
| serine/threonine kinase 31 | gi|13603843 (+2) | 115 kDa | 3 | |
| cell division cycle 5-like protein | gi|22779899 (+3) | 92 kDa | 2 | |
| ASR2B | gi|13517493 (+3) | 100 kDa | 2 | |
| Nuclear-cytoplsimic transport | spermatocytes | spermatids | ||
| RAN GTPase activating protein 1 | gi|148672614 (+5) | 73 kDa | 13 | 2 |
| Ran-specific GTPase-activating protein | gi|153792001 (+2) | 24 kDa | 2 | |
| Ran binding protein 5 | gi|12057236 (+2) | 124 kDa | 6 | 2 |
| Karyopherin (importin) beta 1 | gi|30931411 (+4) | 97 kDa | 4 | 2 |
| exportin-2 | gi|12963737 | 110 kDa | 2 | |
| Transcription, RNA-interaction/ stability, splicing | spermatocytes | spermatids | ||
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | gi|148686462 (+2) | 78 kDa | 4 | |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 | gi|133777033 (+6) | 89 kDa | 2 | |
| piwi-like protein 2 | gi|10946610 | 109 kDa | 5 | |
| paraspeckle component 1 | gi|225543409 (+2) | 59 kDa | 2 | |
| RNA polymerase II largest subunit | gi|2145091 (+2) | 217 kDa | 3 | |
| RNA-binding protein EWS | gi|88853581 (+4) | 69 kDa | 3 | 3 |
| RNA-binding protein 14 | gi|86262142 (+1) | 36 kDa | 7 | 4 |
| TAR DNA-binding protein 43 isoform 1 | gi|21704096 (+4) | 45 kDa | 2 | |
| PC4 and SFRS1-interacting protein | gi|19527168 | 51 kDa | 5 | 6 |
| elongation factor 2 | gi|33859482 (+8) | 19 kDa | 4 | 4 |
| splicing factor, proline- and glutamine-rich | gi|23956214 (+1) | 60 kDa | ||
| splicing factor, arginine/serine-rich 14 | gi|148696857 (+4) | 120 kDa | 2 | |
| splicing factor 3B subunit 1 | gi|15214281 (+1) | 146 kDa | 3 | |
| splicing factor 1 protein | gi|14318588 (+10) | 67 kDa | 6 | 2 |
| splicing factor, arginine/serine-rich 15 | gi|109150409 | 2 | ||
| far upstream element (FUSE)-binding protein 2 | gi|163954948 (+2) | 75 kDa | 7 | 9 |
| far upstream element (FUSE) binding protein 3 | gi|224922832 (+2) | 62 kDa | 2 | |
| fusion, derived from t(12;16) malignant liposarcoma | gi|148685669 (+6) | 98 kDa | 3 | 6 |
| interleukin enhancer-binding factor 3 isoform 1 | gi|111607430 (+7) | 60 kDa | 3 | |
| T-complex protein 1 subunit epsilon | gi|6671702 (+6) | 68 kDa | 6 | 5 |
| nonsense mRNA reducing factor 1 NORF1 | gi|12836885 (+4) | 7 | 2 | |
| THO complex subunit 4 | gi|6755763 (+1) | 27 kDa | 3 | 2 |
| HLA-B-associated transcript 3 | gi|148694699 (+7) | 111 kDa | 2 | |
| polypyrimidine tract binding protein 2 | gi|148680404 (+4) | 68 kDa | 4 | |
| prohibitin-2 | gi|126723336 (+2) | 33 kDa | 2 | |
| Abce1 protein, partial [ | gi|45219736 (+3) | 65 kDa | 2 | |
| Enhancer of mRNA-decapping protein 4 | gi|145566774 (+4) | 152 kDa | 3 | |
| protein strawberry notch homolog 1 | gi|124487087 (+2) | 154 kDa | 3 | |
| Ribonucleoproteins | spermatocytes | spermatids | ||
| heterogeneous nuclear ribonucleoprotein A/B isoform 1 | gi|146260280 (+5) | 95 kDa | 5 | 3 |
| heterogeneous nuclear ribonucleoprotein A1 isoform b | gi|85060507 (+6) | 16 kDa | 6 | |
| heterogeneous nuclear ribonucleoprotein U | gi|148681230 (+6) | 49 kDa | 5 | |
| heterogeneous nuclear ribonucleoprotein H | gi|10946928 (+2) | 123 kDa | 2 | 5 |
| small nuclear ribonucleoprotein N | gi|3142634 (+2) | 25 kDa | 3 | |
| heterogeneous nuclear ribonucleoprotein Q isoform 1 | gi|114145493 (+9) | 70 kDa | 4 | |
| heterogeneous nuclear ribonucleoprotein L | gi|183980004 (+6) | 64 kDa | 2 | |
| heterogeneous nuclear ribonucleoprotein A3 isoform a | gi|31559916 (+11) | 40 kDa | 2 | |
| heterogeneous nuclear ribonucleoprotein r protein, | gi|13435603 (+6) | 67 kDa | 2 | |
| 116 kDa U5 small nuclear ribonucleoprotein component isoform b | gi|158508674 (+5) | 109 kDa | 2 | |
| heterogeneous nuclear ribonucleoprotein D-like | gi|148664250 (+2) | 46 kDa | 2 | |
| Ribosomal proteins | spermatocytes | spermatids | ||
| 60S ribosomal protein L31-like isoform 1 | gi|82898755 (+6) | 14 kDa | 2 | |
| 60S ribosomal protein L21 | gi|31560385 (+13) | 39 kDa | 2 | |
| 60S ribosomal protein L14 | gi|13385472 (+1) | 24 kDa | 2 | |
| 60S ribosomal protein L22 | gi|6677775 | 15 kDa | 2 | |
| 60S ribosomal protein L23-like | gi|407262287 (+2) | 14 kDa | 2 | |
| 40S ribosomal protein S13 | gi|15029927 (+2) | 16 kDa | 2 | |
| 40S ribosomal protein S20 isoform 2 | gi|4506697 | 13 kDa | 2 | |
| 40S ribosomal protein S19 | gi|12963511 (+3) | 16 kDa | 2 | |
| 16S ribosomal protein | gi|200796 (+1) | 87 kDa | 2 | 2 |
| ribosomal protein S26 | gi|1527176 (+3) | 13 kDa | 2 | 2 |
| Ribosomal protein S9 | gi|21594169 (+1) | 23 kDa | 3 | 2 |
| Ribosomal protein S23 | gi|72679974 (+5) | 16 kDa | 2 | |
| La ribonucleoprotein domain family member 1 | gi|147744571 (+3) | 121 kDa | 2 | |
| Cytoskeleton | spermatocytes | spermatids | ||
| lamin-B1 | gi|188219589 (+2) | 67 kDa | 3 | 6 |
| alpha-actinin-4 | gi|11230802 | 105 kDa | 2 | |
| filamin-C | gi|124487139 (+2) | 291 kDa | 10 | |
| filamin-B | gi|145966915 (+2) | 277 kDa | 8 | |
| talin 1 | gi|148670519 (+4) | 270 kDa | 3 | |
| vinculin | gi|148669535 (+3) | 124 kDa | 3 | |
| coiled-coil domain containing 39 | gi|148703084 (+2) | 107 kDa | 2 | |
| kinesin family member 5B | gi|148691088 (+3) | 110 kDa | 4 | |
| protein syndesmos | gi|13385314 (+1) | 23 kDa | 3 | |
| plectin | gi|122065897 (+24) | 534 kDa | 2 | |
| myosin light chain, regulatory B-like | gi|71037403 (+4) | 20 kDa | 3 | |
| Membrane-associated, vesicle trafficking, ER proteins | spermatocytes | spermatids | ||
| acrosin-binding protein isoform 1 precursor | gi|188035922 (+1) | 61 kDa | 2 | |
| GPI-anchored membrane protein 1 | gi|148695758 (+7) | 84 kDa | 4 | 4 |
| ras-related protein Rab-11B | gi|6679583 (+3) | 24 kDa | 5 | 2 |
| ribophorin I | gi|148666824 (+4) | 68 kDa | 2 | |
| ribophorin | gi|1468961 (+4) | 66 kDa | 7 | 2 |
| calreticulin precursor | gi|6680836 (+5) | 48 kDa | 2 | |
| rab7 | gi|1050551 (+1) | 24 kDa | 3 | 2 |
| Programmed cell death 6 interacting protein | gi|20071292 (+8) | 96 kDa | 4 | 2 |
| calmegin, isoform CRA_b | gi|148678956 (+3) | 71 kDa | 3 | 2 |
| calnexin precursor | gi|6671664 (+1) | 67 kDa | 2 | |
| solute carrier family 2, facilitated glucose transporter member 3 | gi|261862282 (+2) | 53 kDa | 3 | 3 |
| transmembrane emp24-like trafficking protein 10 | gi|148670919 (+2) | 26 kDa | 2 | |
| ras-related protein Rab-14 | gi|18390323 (+3) | 24 kDa | 3 | |
| SEC22 vesicle trafficking protein homolog B | gi|14290512 (+1) | 25 kDa | 2 | |
| ERO1-like beta | gi|109730421 (+3) | 54 kDa | 2 | |
| p162 protein | gi|1205976 (+7) | 162 kDa | 2 | |
| zinc finger protein 289 | gi|148695611 (+4) | 58 kDa | 2 | |
| Glycolitic and mitochondria enzymes | spermatocytes | spermatids | ||
| cytochrome c oxidase subunit 6C | gi|16716343 | 8 kDa | 2 | |
| cytochrome b-cl complex subunit 2, mitochondrial precurso | gi|22267442 (+2) | 48 kDa | 2 | |
| cytochrome b-c1 complex subunit 1, mitochondrial precursor | gi|46593021 (+3) | 53 kDa | 3 | |
| cytochrome c oxidase subunit IV | gi|1372988 (+2) | 20 kDa | 3 | |
| dihydrolipoamide S-acetyltransferase precursor | gi|16580128 (+2) | 59 kDa | 3 | |
| trifunctional enzyme subunit alpha, mitochondrial precursor | gi|33859811 (+1) | 83 kDa | 6 | |
| trifunctional enzyme subunit beta, mitochondrial precursor | gi|21704100 (+4) | 51 kDa | 2 | |
| phosphoglycerate mutase 1 | gi|114326546 (+1) | 29 kDa | 2 | |
| L-lactate dehydrogenase A chain isoform 2 | gi|257743039 (+5) | 40 kDa | 3 | 2 |
| L-lactate dehydrogenase B chain | gi|6678674 | 37 kDa | 2 | 2 |
| phosphate carrier protein, mitochondrial precursor | gi|19526818 (+6) | 40 kDa | 2 | |
| fructose-bisphosphate aldolase A isoform 1 precursor | gi|293597567 (+2) | 45 kDa | 5 | |
| aldehyde dehydrogenase 2, mitochondria | gi|148687772 (+5) | 55 kDa | 4 | |
| Me1 protein | gi|13096987 (+6) | 64 kDa | 2 | 2 |
| ATP synthase subunit O, mitochondrial precursor | gi|20070412 (+2) | 23 kDa | 3 | |
| inositol-3-phosphate synthase 1 | gi|12963757 | 61 kDa | 2 | |
| inner membrane protein, mitochondrial | gi|148666538 (+10) | 81 kDa | 2 | |
| creatine kinase B-type | gi|10946574 (+3) | 43 kDa | 3 | |
| acyl-CoA hydrolase | gi|14587839 (+6) | 38 kDa | 2 | |
| N(4)-(beta-N-acetylglucosaminyl)-L-asparaginase isoform 1 precursor | gi|54292135 | 37 kDa | 3 | |
| carnitine O-palmitoyltransferase 2, mitochondrial precursor | gi|162138915 (+3) | 74 kDa | 2 | |
| isocitrate dehydrogenase 3 (NAD+) alpha | gi|148693872 (+5) | 40 kDa | 2 | |
| enoyl-CoA delta isomerase 1, mitochondrial precursor | gi|31981810 (+1) | 32 kDa | 2 | |
| coiled-coil-helix-coiled-coil-helix domain containing 3 | gi|148681756 (+2) | 23 kDa | 2 | |
| Other enzymes | spermatocytes | spermatids | ||
| GMP synthase [glutamine-hydrolyzing] | gi|85861218 (+4) | 77 kDa | 2 | |
| tripeptidyl peptidase II | gi|148664483 (+4) | 139 kDa | 2 | |
| Aspartyl-tRNA synthetase | gi|14250408 (+6) | 57 kDa | 2 | |
| triosephosphate isomerase, partial | gi|1864018 (+5) | 23 kDa | 4 | |
| retinol dehydrogenase 11 precursor | gi|19482172 (+3) | 35 kDa | 2 | |
| hydroxysteroid (17-beta) dehydrogenase 4 | gi|148677986 (+3) | 33 kDa | 3 | |
| Immunoglobulins | spermatocytes | spermatids | ||
| Ig heavy chain V region TE32 | gi|110285 (+14) | 13 kDa | 2 | 2 |
| kappa-Ig light chain (111 AA) | gi|930228 | 12 kDa | 9 | |
| Unknown functions in testis | spermatocytes | spermatids | ||
| ataxin 2-like | gi|148685438 | 113 kDa | 10 | 4 |
| testis specific 10 | gi|148682582 (+2) | 80 kDa | 4 | |
| neuroleukin | gi|200065 (+8) | 63 kDa | 2 | 2 |
| myelin expression factor 2 isoform 1 | gi|244790087 (+4) | 63 kDa | 2 | |
| platelet-activating factor acetylhydrolase IB subunit gamma | gi|6679201 | 26 kDa | 2 | |
| interleukin enhancer-binding factor 2 | gi|13385872 (+2) | 43 kDa | 2 | |
| tetratricopeptide repeat protein 21B | gi|114158711 (+1) | 151 kDa | 2 | |
| annexin A3 | gi|148688409 (+6) | 36 kDa | 2 | |
| high density lipoprotein (HDL) binding protein | gi|148708002 (+5) | 144 kDa | 2 | |
the number showing in the bracket following “+” means how many more accession number also refer to this target
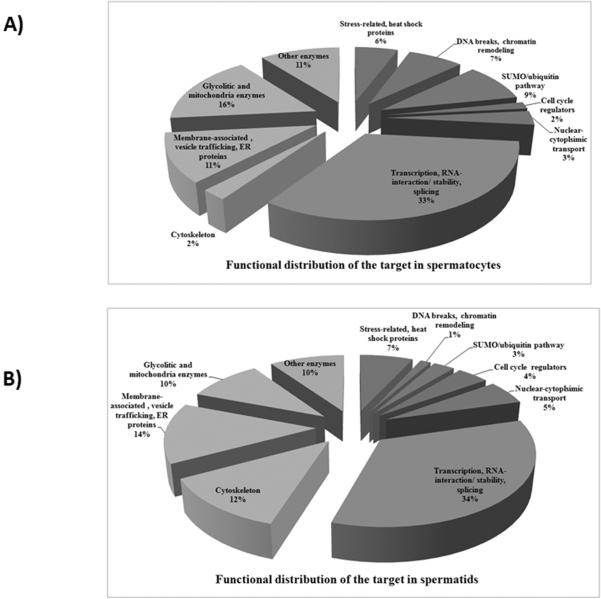
Figure 2.
Functional distribution of SUMO targets in mouse spermatocytes (A) and spermatids (B). The percentage of sumoylated proteins from a certain functional group out of 100% of the identified sumoylated proteins is shown.
Glycolytic and mitochondrial enzymes were found to be sumoylated in both spermatocytes (16%) and spermatids (10%), together with other enzymes (10–11%). Several proteins involved in ubiquitination, including ubiquitin-activating and ubiquitin-conjugating enzymes, ubiquitin hydrolases, and proteasome subunits were identified as SUMO targets in spermatocytes (9%) and in spermatids (3%) (Table 1, Fig. 2).
Stress-related and heat shock proteins represented 6–7% of sumoylated targets in spermatocytes and spermatids. Membrane-associated, vesicle trafficking, and ER proteins represented 11% of sumoylated proteins in spermatocytes and 14% in spermatids. This group included Rab 7 and Rab 11, calreticulin and acrosin-binding protein precursors, and the testis-specific ER protein calmegin (Table 1, Fig. 2).
Cytoskeletal proteins represented only 2% of spermatocyte but 12% of spermatid SUMO targets. Conversely, proteins involved in DNA-break repair and chromatin remodeling were primarily sumoylated in spermatocytes (7%) but not in spermatids (1%). Importantly, this group included several proteins implicated in the regulation of chromatin remodeling during meiosis, such as mediator of DNA damage checkpoint protein 1 (MDC1), SWI/SNF-related regulator of chromatin 4 and 5 (SMARCA4 and 5), and poly [ADP-ribose] polymerase 1 (PARP1) (Table 1, Fig. 2).
Proteins that mediate nuclear-cytoplasmic transport comprised 3% of the spermatocyte and 5% of the spermatid sumoylome. Importantly, the major known sumoylated target in cells, Ran GTPase activating protein 1 (RanGAP1), was identified in both spermatocyte and spermatid fractions. This finding serves as a positive control for the specificity of SUMO target identification. Other proteins in this group included Ran binding protein 5, importin beta, and exportin 2, which are known to interact with RanGAP1(Roscioli et al. 2012).
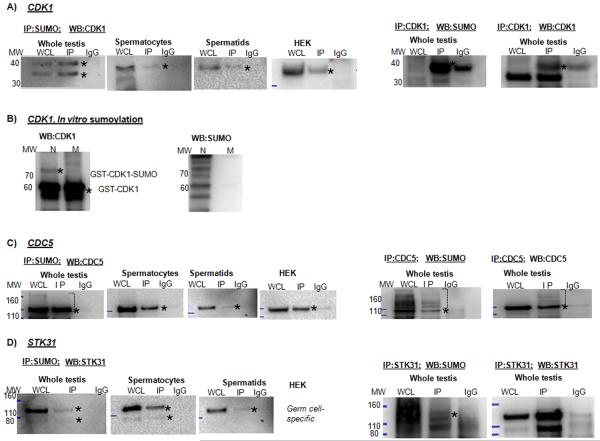
Cell cycle regulators represented 2% and 4% of sumoylated proteins in spermatocytes and spermatids, respectively. These important regulators of meiosis and mitosis include CDK1 (CDC2), testis-specific serine/threonine kinase 31 (STK31) and cell division cycle 5-like protein (CDC5) (Table 1, Fig. 2).
III. Confirmation of possible sumoylated targets
GPS-SUMO, a tool for the prediction of sumoylation sites and SUMO-interaction motifs (SIMs), was used to identify consensus and non-consensus sequences for possible sumoylation within the amino acid sequences of the proteins (Zhao et al. 2014). Because approximately 40% of proteins that are sumoylated are not sumoylated within the consensus sequence, the prediction algorithm of GPS-SUMO is based on the analysis of 983 manually collected consensus and non-consensus sumoylation sites in 545 proteins and 137 known SIMs in 80 proteins (Zhao et al. 2014). Several proteins with important roles during spermatogenesis (displaying an infertility phenotype upon inactivation or/and germ cell-specific proteins) were analyzed for the presence of sumoylation sites, and several of those with multiple and/or conserved sites were chosen for further characterization. These proteins included MDC1, KAP1, MILI, DDX4, CDK1, CDC5, STK31, TDP-43 and the largest subunit of RNAP II. Co-IP assays performed with anti-SUMO1 and anti-target protein antibodies alongside their negative controls were followed by Western blot analyses with antibodies against the target proteins or SUMO.
III.1. RNAP II
RNA polymerase II (RNAP II) is a master regulator of transcription in both germ and somatic cells. RNAPII was identified as SUMO-interacting protein specifically in the spermatocyte fraction mass spectrometry screen (Table 1). In lysates from whole testis lysate, purified spermatocytes, spermatids and human embryonic kidney (HEK) cells, an isoform of RNAP II was specifically identified with an anti-SUMO antibody followed by Western blotting with an antibody against the largest subunit of RNAP II, suggesting that this is a sumoylated isoform (Fig. 3A). IP with an anti-RNAP II antibody did not successfully enrich for sumoylated isoforms of the proteins (not shown), which was likely due to the less efficient recognition by the antibody of the sumoylated isoform of the protein compared with the non-sumoylated form. Bioinformatics analysis revealed the presence of only non-consensus sumoylation sites in the amino acid sequence of RNAP II. However, the two sequences were evolutionary conserved between mouse and human (Supplementary Table 2).
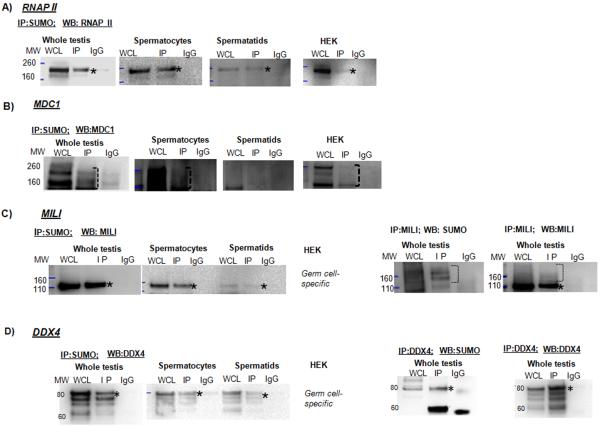
Figure 3.
Co-IP analysis of SUMO and RNAP II (A), MDC1 (B), MILI (C), and DDX4 (D). Whole-cell lysate (WCL), negative control (IgG) and IP fractions are shown. The migrating positions of the molecular weight (MW) markers are indicated.
(A) Co-IP analysis of SUMO and RNAP II. In lysates prepared using denaturing buffer from whole testis, purified spermatocytes, spermatids, and Human Embryonic Kidney (HEK)-293 cells, two isoforms of RNAP II were identified. The isoform with a higher molecular weight (asterisk) was identified by IP with an anti-SUMO antibody followed by Western blotting with an anti-RNAP II antibody.
(B) Using whole testis, purified spermatocytes, and Human Embryonic Kidney (HEK)-293 cells lysate prepared using non-denaturing buffer, IP with an anti-SUMO antibody followed by Western blotting with an MDC1 antibody detected multiple isoforms of MDC1 (bracket). There was no specific signal identified in the spermatid fraction,.
(C) IP using anti-SUMO or anti-MILI antibodies followed by Western blotting with either anti-MILI or anti-SUMO antibodies. Using spermatocytes, spermatids and whole testis lysate prepared using non-denaturing buffer, IP with an anti-SUMO antibody followed by Western blotting with an anti-MILI antibody identified a non-covalent interaction of the protein with SUMO or sumoylated proteins (a band approximately 110 kDa; asterisk). The signal in spermatids was at a low level. Sumoylated conjugates of higher molecular weight were precipitated with an anti-MILI antibody followed by Western blotting with either the anti-SUMO or anti-MILI antibody.
(D) IP using SUMO or DDX4 antibodies followed by Western blotting with either DDX4 or SUMO antibodies. Several isoforms of the protein could be detected on the Western blot using an anti-DDX4 antibody and non-denaturing lysis buffer. The largest isoform (approximately 80 kDa, asterisk) could be a sumoylated form of the protein, as it was specifically identified by reciprocal co-IP using both SUMO and DDX4 antibodies.
III.2. MDC1
MDC1 is an important regulator of the DNA damage response. Male-specific infertility in Mdc1−/− mice is due to meiotic arrest (Lou et al. 2006). MDC1 is an essential factor for establishing sex chromosome-wide silencing in the pachytene stage (Kunin et al. 2010, Ichijima et al. 2011). MDC1 was specifically identified in the spermatocyte fraction in our mass-spectrometry screen (Table 1). Several MDC1 isoforms in the range of 160–260 kDa that may correspond to splice isoforms of the protein were pulled down using co-IP with anti-SUMO antibody followed by MDC1 Western blot from the whole testis, purified spermatocyte but not spermatid fraction lysate (Fig. 3B). Sumoylation of MDC1 in somatic cells was previously reported (Yin et al. 2012). Bioinformatics analysis revealed the presence of several consensus sumoylation sites in both the mouse and human amino acid sequences of MDC1 (Supplementary Table 2).
III.3. MILI
MILI, a mammalian member of the Piwi gene family, binds to piRNAs. Spermatogenesis in mili-null mice is blocked at early prophase of the first meiosis, and the mice are sterile (Kuramochi-Miyagawa et al. 2004). MILI was specifically identified in the spermatocyte fraction in the mass-spectrometry screen (Table 1). IP with an anti-SUMO antibody followed by Western blotting with an anti-MILI antibody identified an apparent non-covalent interaction of the protein with SUMO or sumoylated proteins in spermatocytes and whole testis lysate (a band of approximately 110 kDa, Fig. 3C). The signal in spermatid fraction was very weak suggesting either an absence or a low level of SUMO and MILI interaction in these cells. Higher molecular weight (and presumably sumoylated) isoforms of MILI precipitated better with an anti-MILI antibody than with the anti-SUMO antibody followed by Western blotting with either the anti-SUMO or anti-MILI antibody. Overall, these results suggest that there is both covalent and non-covalent modification of MILI by SUMO. MILI is a germ cell specific protein, and therefore somatic data are not shown. Bioinformatics analysis revealed the presence of several consensus sumoylation sites in both the mouse and human amino acid sequences of the protein (Supplementary Table 2).
III.4. DDX4
DDX4 is the mouse VASA homologue (MVH) that is expressed exclusively in germ cells. It interacts with MILI to regulate microRNA-mediated RNA silencing (Kuramochi-Miyagawa et al. 2004). DDX4 is required for the development of male germ cells. Male mice that are homozygous for a targeted mutation of Mvh produce no sperm in the testes, with spermatogenic arrest at early meiosis in a manner similar to MILI-deficient mice (Tanaka et al. 2000). Several isoforms of the protein ranging from 60–80 kDa were detected by Western blotting using an anti-DDX4 antibody in spermatocytes, spermatids and whole testis lysate. These proteins may be the result of alternative splicing (Luo et al. 2013), Fig. 3D). The largest isoform (approximately 80 kDa) may be a sumoylated form of the protein, as this isoform was specifically identified by reciprocal co-IP using both SUMO and DDX4 antibodies. Although DDX4 was identified in spermatocyte fraction in the mass-spectrometry screen (Table 1), a more sensitive approach of Western blotting supported sumoylation of DDX4 in both spermatocytes and spermatids. DDX4 is a germ-cell specific protein, and therefore somatic data are not shown. Bioinformatics analyses revealed the presence of several consensus sumoylation sites in both the mouse and human amino acid sequences of DDX4, supporting possible sumoylation (Supplementary Table 2).
III.5. KAP1
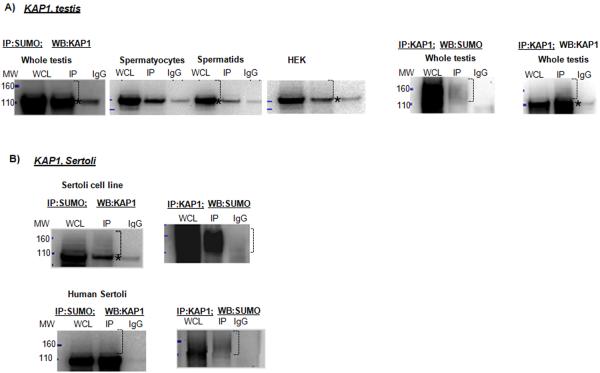
KRAB domain-associated protein 1 (KAP1, TIF1β, TRIM28) is a transcriptional repressor known to play essential roles in chromatin remodeling in early embryonic development and spermatogenesis. During spermatogenesis, KAP1 is preferentially associated with the heterochromatin structures of spermatocytes, spermatids and Sertoli cells. KAPI was identified in both the spermatocyte and spermatid fractions in the mass-spectrometry screen (Table 1). IP with an anti-SUMO antibody followed by KAP1 Western blot analysis identified a possible non-covalent interaction of the protein with SUMO (a band at approximately 100 kDa, Fig. 4A) in the whole testis, spermatocyte, spermatid, and HEK cell lysate. Higher molecular weight SUMO-conjugates were clearly detected by IP with an anti-KAP1 antibody followed by Western blotting with either an anti-SUMO or anti-KAP1 antibody (Fig. 4A). KAP1 is an important protein in Sertoli cells. Therefore, to further evaluate the possible sumoylation of KAP1 in those cells, we also employed primary human and transformed mouse Sertoli cell lines. In all Sertoli cells, co-IP analyses revealed possible covalent and non-covalent interaction of KAP1 with SUMO (Fig. 4B, human and mouse Sertoli cell lines are shown). These results suggest that Sertoli cell lines can be used to a certain degree to study the regulation of sumoylation in Sertoli cells. Sumoylation of KAP1 in somatic cells was previously reported (Li et al. 2007, Campbell & Izumiya 2012). Bioinformatics analysis revealed the presence of several consensus sumoylation sites in both the mouse and human amino acid sequences of the protein (Supplementary Table 2).
Figure 4.
Co-IP analysis of SUMO and KAP1. IP using non-denaturing lysis buffer with anti-SUMO antibody followed by KAP1 Western blot analysis identified a possible non-covalent interaction of the protein with SUMO (a band approximately 100 kDa, asterisk). Higher molecular weight SUMO-conjugates were clearly detected by IP with an anti-KAP1 antibody followed by Western blotting with either an anti-SUMO or anti-KAP1 antibody (bracket). In a similar manner, co-IP analyses of KAP1 and SUMO in Sertoli cells and cell lines revealed possible covalent and non-covalent interactions of KAP1 with SUMO.
III.6. CDK1
CDK1 (CDC2) is a crucial and indispensable regulator of both mitosis and meiosis (Diril et al. 2012). CDK1 was identified in both the spermatocyte and spermatid fractions in the mass-spectrometry screen (Table 1). Lysates from the whole testis and purified spermatocytes and spermatids were obtained, and two isoforms of CDK1 were identified in the SUMO pull-down from the whole testis and spermatocytes, suggesting both the covalent and non-covalent interaction of CDK1 with SUMO (Fig. 5A). Only one isoform was observed at the detectable level in the spermatid and HEK cell lysate. The higher-molecular weight isoform was also highly enriched after IP with an anti-CDK1 antibody followed by Western blotting with an anti-SUMO antibody (Fig. 5A). Some background signal was detected in the negative control when highly concentrated lysates were used for the CDK1 IP analysis (whole testis). However, the background was significantly lower than the specific signal and was not detected upon IP with anti-SUMO antibodies. To confirm the possible sumoylation of CDK1 and given the importance of the protein in both germ and somatic cells, an in vitro sumoylation reaction was performed with a commercially available recombinant GST-CDK1 protein, sumoylation enzymes (E1, E2), and either normal SUMO or a mutant SUMO incapable of forming an isopeptide bond (Fig. 5B). Western blot analysis with an anti-CDK1 antibody revealed the presence of a sumoylated CDK1 band above the non-modified GST-CDK1 when using the normal (N) but not the mutant (M) SUMO isoform (Fig. 5B). When detected with an anti-SUMO antibody, multiple bands were observed in the sample with normal SUMO, most likely corresponding to sumoylated E1 and E2 in addition to sumoylated CDK1. However, these bands were not observed in the sample with the mutant SUMO isoform. Bioinformatics analysis revealed the presence of the consensus sumoylation site in the amino acid sequence of the mouse but not the human CDK1 (Supplementary Table 2). However, the alignment of the two sequences revealed a difference in only one amino acid, with a possible target lysine still present at the same position. We examined whether another important cell cycle regulator, CDK2 (not identified by our screen), contains a consensus sequence for sumoylation and detected no such sequence in CDK2 (not shown).
Figure 5.
Co-IP analysis of SUMO and CDK1 (A), CDC5 (C), and STK31 (D) and the in vitro sumoylation analysis of CDK1 (B).
(A) Co-IP analysis of SUMO and CDK1. Lysates prepared using denaturing lysis buffer from whole testis and purified spermatocytes were obtained, and two isoforms of CDK1 were identified in the SUMO pulldown, suggesting both the covalent and non-covalent interaction of CDK1 with SUMO. Only one isoform was detectable in spermatids and HEK cells. The higher-molecular weight isoform (asterisk) was also highly enriched after IP with an anti-CDK1 antibody followed by Western blotting with an anti-SUMO antibody.
(B) To confirm the possible sumoylation of CDK1, an in vitro sumoylation reaction was performed with a recombinant GST- CDK1 protein, sumoylation enzymes (E1, E2), and either normal (N) or a mutant (M) SUMO incapable of forming an isopeptide bond. Western blot analysis with an anti-CDK1 antibody revealed the presence of a sumoylated CDK1 band above the non-modified GST-CDK1 when using the normal (N) but not mutant (M) SUMO isoform. When detecting with an anti-SUMO antibody, multiple bands were observed only in the sample with normal SUMO, most likely corresponding to sumoylated E1 and E2 in addition to sumoylated CDK1.
(C) Reciprocal co-IP using non-denaturing lysis buffer, anti-SUMO and anti-CDC5 antibodies support both covalent (bracket) and non-covalent interactions of CDC5 with SUMO (asterisk).
(D) Co-IP analysis using non-denaturing lysis buffer, anti-SUMO and anti-STK31 antibodies supported possible non-covalent and covalent interactions of StkTK31 with SUMO. The asterisk indicates a possible sumoylated isoform found above the non-sumoylated isoform of STK31.
III.7. CDC5
CDC5 is a DNA-binding protein involved in cell cycle control. Using lysates from the whole testis, spermatocytes, spermatids, and HEK cells, reciprocal co-IP using anti-SUMO and anti-CDC5 antibodies supported the mostly non-covalent interactions of CDC5 with SUMO (Fig. 5C, a band approximately 110 kDa) but also some weak bands of higher molecular weight, which can correspond to sumoylated isoforms of the protein (brackets). Although CDC5 was identified in the spermatide fraction in the mass-spectrometry screen, Western blotting supported interaction between CDC5 and SUMO in both spermatocytes and spermatids. Bioinformatics analysis revealed the presence of two consensus sumoylation sites conserved between mouse and human in the amino acid sequences of CDC5 (Supplementary Table 2).
III.8. STK31
StkTK31 is a germ cell specific protein kinase. StkTK31 was identified in both the spermatocyte and spermatid fractions in the mass-spectrometry screen (Table 1). Co-IP analysis with anti-SUMO and anti-STK31 antibodies using the whole testis, spermatocyte and spermatid lysates supported mostly covalent (Fig. 5D, a band at approximately 110 kDa) and some non-covalent (a band just below 80 kDa in some fractions) interactions of STK31 with SUMO (Fig. 5D). Although STK31 was only identified in the spermatocyte fraction in the mass-spectrometry screen, Western blotting supported possible interaction between CDC5 and SUMO in both spermatocytes and spermatids. STK31 is a germ-cell specific protein, and somatic data are not shown. Bioinformatics analysis revealed the presence of multiple conserved consensus sumoylation sites in the amino acid sequences of STK31 in mouse and human (Supplementary Table 2).
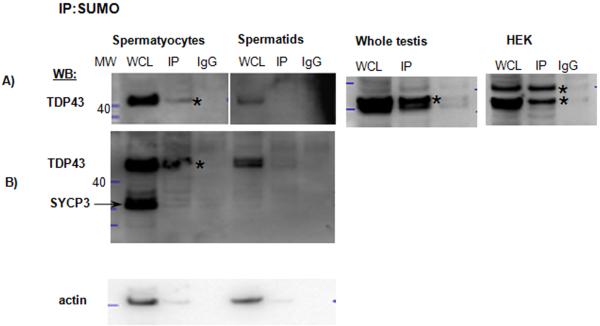
III.9. TDP-43
TDP-43 is an evolutionarily conserved, ubiquitously expressed DNA/RNA-binding protein. In testis, it binds to the promoter of the testis-specific mouse Acrv1 gene in spermatocytes and spermatids, but ACRV1 is expressed exclusively in spermatids. Mutations in the TDP-43 promoter-binding motifs lead to the premature transcription of Acrv1 in spermatocytes. TDP-43 may be involved in pausing RNAPII at the Acrv1 promoter in spermatocytes (Lalmansingh et al. 2011). One mechanism regulating the different activity of TDP-43 in spermatocytes and spermatids could be a posttranslational modification. Our mass spectrometry screen identified TDP-43 as SUMO target in spermatocytes but not in the spermatid fraction (Table 1). Notably, IP with an anti-SUMO antibody followed by TDP-43 Western blot analysis of the whole testis and purified germ cell fractions (Fig. 6A) confirmed that SUMO and TDP-43 interact specifically in spermatocytes but not in spermatids (Fig. 6A and B; two membranes with increasing amounts of protein are shown). The purity of the fractions was confirmed using an anti-SYCP3 antibody showing a prominent band specifically in the spermatocyte fraction (Fig. 6B, SYCP3). Interestingly, two bands were identified using IP in HEK cells with an anti-SUMO antibody followed by TDP-43 Western blot analysis, suggesting both covalent and non-covalent interactions between TDP-43 and SUMO. Bioinformatics analysis revealed the presence of a conserved non-consensus sequence at the same residues in the mouse and human proteins (Supplementary Table 2).
Figure 6.
Co-IP analysis of SUMO and TDP-43. IP using non-denaturing lysis buffer with anti-SUMO antibody followed by TDP-43 Western blot analysis identified a prominent specific signal in the whole testis and spermatocyte but not in the spermatid fractions (A and B show two different membranes with increasing amount of proteins). The same membrane was re-probed with anti-SYCP3 antibody (SYCP3) to confirm the purity of the spermatocyte and spermatid fractions and with an actin antibody to show a comparable level of the whole cell lysates used for the IP. Two bands were identified in HEK cells, suggesting both covalent and non-covalent interactions between TDP-43 and SUMO.
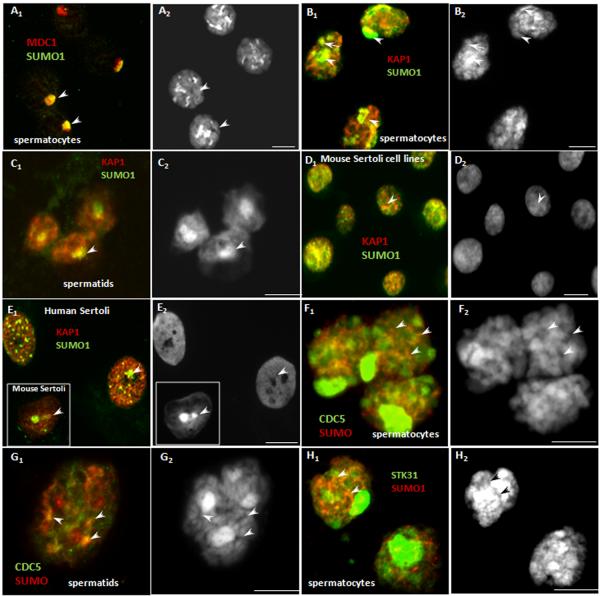
IV. Colocalization studies
Partial colocalization of SUMO with its putative targets (orange signal) supported their possible sumoylation. Several targets showed a certain degree of colocalization between SUMO in the large heterochromatic regions of spermatocytes (MDC1 in the XY body (Fig. 7 A1 and A2, arrowheads) and KAP1 in the centromeric heterochromatin and partially in the XY body of spermatocytes (arrow and arrowheads, respectively, Fig. 7B1 and B2) and chromocenters of round spermatids (Fig. 7C1 and C2, arrowheads). KAP1 also colocalized with SUMO in certain areas of human and mouse Sertoli cells as well as in mouse Sertoli cell lines (Fig. 7, D1, D2; E1, E2, in insert: mouse Sertoli). The degree of colocalization varied between the cells, suggesting either a cell-cycle or stage-dependent behavior. Several targets of SUMO involved in transcriptional regulation and microRNA biogenesis (e.g., CDC5 (F1, F2, G1, G2), STK31 (H1, H2)) showed some overlap with SUMO that resulted in sparse orange signals detectable primarily in the DAPI-poor areas of spermatocytes. These areas may correspond to intrachromosomal territories (Branco & Pombo 2006).
Figure 7.
Immunofluorescent localization of SUMO and its putative targets in testicular cells. Targets, color-coding and cell type are indicated for each image. A mouse monoclonal anti-SUMO1/GMP antibody was used in (A1), and a rabbit monoclonal anti-SUMO1 was used in (B–H). Nuclei are stained by DAPI (blue). For all images, immunofluorescent staining is shown alongside the corresponding DAPI images to demonstrate the chromatin structure. Scale bar is 10 μm. SUMO colocalized with MDC1 in the XY body (Fig. 7 A1 and A2, arrowheads) and KAP1 in the centromeric heterochromatin and partially in the XY body of spermatocytes (Fig. 7B1 and B2, arrowheads and arrow, respectively) and chromocenters of round spermatids (Fig. 7C1 and C2, arrowheads). KAP1 also colocalized with SUMO in certain areas of human and mouse Sertoli cells as well as in mouse Sertoli cell lines (D1, D2; E1, E2, an insert: mouse Sertoli). CDC5 (F1, F2,G1, G2), and STK31 (H1, H2) were detectable primarily in the DAPI-poor areas of cells that may correspond to intrachromasomal domains (arrows).
Discussion
In this study, immunoprecipitation and Western blot analysis with an anti-SUMO1 antibody were used to identify SUMO targets in testicular cells. Our previous study showed similar localization and Western blot patterns of SUMO1 and SUMO2/3 in testicular cells and sperm. Together with an absence of defects in spermatogenesis in SUMO1-knockout mice (Zhang et al. 2008), these data suggest that the functions and targets of SUMO1 and SUMO2/3 overlap in testicular cells. In other cell types, certain proteins are preferentially modified by either SUMO1 or 2/3. These data suggest that the presence of specific SUMO1 and SUMO2/3 targets during spermatogenesis cannot be excluded.
Our results suggest the role of sumoylation as a major player in the regulation of transcription, stress responses, the regulation of major enzymatic pathways, nuclear-cytoplasmic transport, cell cycle control, acrosome biogenesis and other functions in spermatogenesis. Interestingly, proteins involved in ubiquitination and sumoylation, DNA repair, and chromatin remodeling are highly sumoylated in spermatocytes, whereas cytoskeleton proteins are highly modified by SUMO in spermatids. It is possible that these proteins are more dynamically regulated specific cell types, as consistent with their functions.
Several proteins identified as SUMO targets in this study were previously found to be sumoylated in somatic cells (studies from other groups) or in human sperm (a study from our group). These proteins included RanGAP1, MDC1, KAP1, heat shock proteins, heteronucleoproteins, and several splicing factors (Mahajan et al. 1998, Vassileva & Matunis 2004, Li et al. 2007, Matafora et al. 2009, Li et al. 2010, Yin et al. 2012, Vigodner et al. 2013). Although previous studies in mammalian cells did not identify RNAP II as a SUMO target, studies in yeast identified the largest subunit of RNAP II as being sumoylated (Chen et al. 2009). In a similar manner, CDK1 was identified as a SUMO target in Drosophila (Nie et al. 2009). These previous data strongly support our findings and the specificity of our identification. Our results are further supported by the fact that all of the studied targets, with the exception of RNAP II and TDP-43, contain one or more consensus sumoylation sequence/s, and many of these sequences are evolutionarily conserved (Supplementary table 2). RNAP II and TDP-43 have non-consensus sequences, but those sequences are evolutionarily conserved.
Immunoprecipitation of endogenous proteins is challenging because sometimes only a small fraction of certain proteins can be sumoylated at a given time. In some experiments, possible sumoylated isoforms were precipitated with and anti-SUMO antibody but not with the antibody against the target protein (e.g., DDX4, Fig. 3D). These results are probably due to a lower affinity of specific antibodies to the sumoylated form of the protein. For other targets (e.g., MILI, CDC5, Fig. 3C and 5C), sumoylated isoforms were only precipitated with the antibody against the target protein and not with the SUMO antibody. These results can likely be explained by a very large number of sumoylated proteins in cells and a limited ability of the anti-SUMO antibody to precipitate 100% of the sumoylated proteins, particularly those with a very low degree of sumoylation.
Localization studies support the mass spectrometry results regarding a potentially diverse role of sumoylation in germ cells (Fig. 7). Several targets show a partial overlap with SUMO (orange signal). For example, KAP1 is preferentially associated with the heterochromatin structures of spermatocytes, spermatids and Sertoli cells. Interestingly, in somatic cells KAP1 can catalyze its own sumoylation and transcriptional repression in a phosphorylation-dependent manner (Li et al. 2007, Li et al. 2010). Germ cell expression of KAP1 is required for spermatogenesis in the mouse. However, the shedding of immature germ cells in mutant mice was attributed to impaired paracrine interactions between germ and Sertoli cells (Weber et al. 2002, Herzog et al. 2011). Therefore, the role and regulation of KAP1 sumoylation in the heterochromatic region of germ and Sertoli cells should be further examined.
MDC1, which colocalizes with SUMO in the XY body, was recently implicated in spreading heterochromatin over the sex chromosome during MSCI (Ichijima et al. 2011). In somatic cells, MDC1 is sumoylated in response to the formation of double-stranded DNA breaks (Yin et al. 2012). SUMO and several other proteins involved in heterochromatin formation were absent from the sex body of Mdc1-null spermatocytes. Based on these results, the authors of this early study concluded that MDC1 is upstream of SUMO during MSCI. Our results complicated this interpretation somewhat because the sumoylated target itself is not present in Mdc1-null spermatocytes. MDC1 also plays a role in DNA double-stranded break repair and can be SUMO modified in response to ionizing radiation. Sumoylated MDC1 then recruits the ubiquitin ligase RNF4, which mediates ubiquitination at the DNA damage site (Yin et al. 2012). Our previous study showed that SUMO is also localized to the DNA breaks in germ cells (Shrivastava et al. 2010). These data, together with the results of this study, suggest that MDC1 can be sumoylated at the meiotic DNA breaks. Whether this modification activates ubiquitination remains to be determined, but our data support a close crosstalk between ubiquitination and sumoylation. Specifically, several ubiquitin-activating and ubiquitin-conjugating enzymes as well as ubiquitin hydrolyses were identified as SUMO targets (Table 1). Furthermore, similar to previous results from our group obtained for human sperm, ubiquitin was identified in SUMO pulldowns. These results suggest that some proteins are simultaneously modified by sumoylation and ubiquitination (Vigodner et al. 2013).
Several targets implicated in the regulation of transcription and/or microRNA biogenesis were found to interact with SUMO (Fig. 3). Interestingly, MILI was mostly sumoylated in spermatocytes. These data are consistent with a specific role for this protein during meiosis.
TDP-43 is an ubiquitously expressed transcription factor that is highly conserved through evolution (Lalmansingh et al. 2011). There has been an increasing interest in this protein since mislocalized TDP-43 was found in the intracellular ubiquitinated inclusions in the brains of patients with frontotemporal lobar degeneration with ubiquitin-positive inclusions, amyotrophic lateral sclerosis, and Alzheimer disease (Neumann et al. 2006, Lalmansingh et al. 2011). In the testis, TDP-43 regulates the spermatid-specific transcription of Acrv1, and mutations in the Acrv1 promoter-binding motifs of TDP-43 cause premature expression of Acrv1 in spermatocytes. Surprisingly, TDP-43 is also found at the Acrv1 promoter in spermatocytes, where it was suggested to regulate RNAP II pausing by an unknown mechanism. Our results revealed a striking difference in the sumoylation or SUMO-interaction of TDP-43 in spermatocytes and spermatids, suggesting that these interactions can contribute to RNAP II pausing or other cell-specific mechanisms in spermatocytes. Further studies will characterize how sumoylation regulates the functions of TDP-43 in germ cells and other tissues.
An interesting finding of this study was that several kinases (CDK1, CDC5 and STK31) were identified as being sumoylated. CDK1 (CDC2) is a crucial and indispensable regulator of both mitosis and meiosis (Diril et al. 2012). Although CDK1 was not reported to be sumoylated in somatic cells, it was identified as a target of sumoylation in the Drosophila embryo, supporting our finding (Nie et al. 2009). Further studies will need to uncover how the sumoylation of CDK1 affects mitotic and meiotic progression.
STK31 was first identified as a germ cell-specific kinase (Wang et al. 2001) and was reported to interact with MIWI, suggesting a role in miRNA biogenesis and spermatogenesis (Bao et al. 2012). However, genetic studies utilizing mouse models have shown that STK31 is dispensable for spermatogenesis and oogenesis. Nevertheless, human STK31 was reported to be expressed in gastrointestinal cancers, including esophageal, gastric, colon and colorectal cancers (Yokoe et al. 2008, Fok et al. 2012). The knockdown of Stk31 in colon cancer cells promotes cell differentiation and suppresses tumorigenicity (Fok et al. 2012). The role and regulation of STK31by sumoylation in tumorigenesis remain to be characterized.
CDC5 is a DNA-binding protein involved in cell cycle control. Similar to MILI and DDX4, CDC5 is associated with the production of microRNAs through interactions with their gene promoters and RNAP II (Zhang et al. 2013). In yeast, CDC5 is also involved in the regulation of the SUMO pathway and modulates the maintenance and dissolution of cohesion at centromeres (Baldwin et al. 2009, Attner et al. 2013). The functions of CDC5 and its regulation by posttranslation modifications in mammalian spermatogenesis are not yet known.
Although the present data focused on proteins uniquely identified in the antibody fraction and not in the control fraction, some proteins, including those with an important role in spermatogenesis (such as HSP 70–2, phosphorylated H2AX, other histones, topoisomerase 2 alpha, and PIWI-like protein 1), were identified in both fractions (data not shown). The observation that these proteins bind to beads in a non-specific manner does not exclude the possibility that sumoylation or interaction with SUMO occurs. Indeed, TOP2A was identified as a specific target of sumoylation in our previous work (Shrivastava et al. 2010). Further studies will need to be conducted to test the possible sumoylation of these proteins. Furthermore, there may be additional proteins in germ cells that are modified by sumoylation but that were below the detection level of the technique used in this study.
Interestingly, most of the sumoylated proteins analyzed in this study also non-covalently interact with SUMO or sumoylated proteins. Bioinformatics analysis revealed that only four proteins (MDC1, RNAP II, STK31, TDP-43) contained a sumo-interacting motif (SIM, not shown). These data suggest that a greater number of proteins can interact with SUMO or sumoylated proteins non-covalently, regardless of the presence of a SIM.
In conclusion, this study identified and confirmed the sumoylation of several novel, previously uncharacterized SUMO targets, such as CDK1, RNAP II, CDC5, MILI, DDX4, TDP-43 and STK31. Furthermore, several proteins that were previously identified as SUMO targets in somatic cells (e.g., KAP1, MDC1) were identified as SUMO targets in germ cells. Many of these proteins have unique roles in spermatogenesis, particularly during meiotic progression. This research opens a novel avenue for further studies of SUMO at the level of individual targets. If the sumoylation sites of the selected proteins are not identified, site-directed mutagenesis can then be employed to mutate candidate lysine residues to arginine to determine whether this substitution causes the disappearance of the sumoylated isoform(s). New approaches to identify sumoylated sites with mass spectrometry are under development but require further validation (Hsiao et al. 2009). After identification of the acceptor lysine residue(s), an attempt can be made to produce specific antibodies against the sumoylated form of the protein that can then be used for localization and interaction studies. Analysis of the functional consequences of the mutations in the sumoylated sites of the identified proteins, as well as other aspects of impaired sumoylation in germ cells, can be addressed both in vitro and in vivo.
Supplementary Material
Supplementary Figure 1. A histogram of the whole testis single-cell suspension prior to the STA-PUT procedure showed peaks of the haploid, diploid, and tetraploid cell populations (A). Representative images of fractions enriched for tetraploid and haploid cells after STA-PUT separation are shown. (B). Immunofluorescence experiments were carried out to examine the fraction purity and presence of sumoylated proteins upon completion of the prolonged separation procedure. SUMO was still localized to the centromeric heterochromatin (arrows) of spermatocytes and round spermatids, as well as to the sex body of spermatocytes (arrowheads). Scale bar is 10 μm.
Supplementary Figure 2. No significant difference was found between the results of a co-IP performed using denaturating and non-denaturating lysis buffers.
Acknowledgment
This study was supported by the NIH, NICHD, and Academic Research Enhancement Award 1R15HD067944-01A1 (MV, PI). Undergraduate student research was supported by Selma and Jacques H. Mitrani Foundation.
Grant support: This study was supported by the NIH, NICHD, and Academic Research Enhancement Award 1R15HD067944-01A1 (MV, PI).
Footnotes
Conflict of interests: The authors declare no conflict of interest.
References
- Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Matic I, Vertegaal AC. Identification of SUMO target proteins by quantitative proteomics. Methods Mol Biol. 2009;497:19–31. doi: 10.1007/978-1-59745-566-4_2. [DOI] [PubMed] [Google Scholar]
- Attner MA, Miller MP, Ee LS, Elkin SK, Amon A. Polo kinase Cdc5 is a central regulator of meiosis I. Proc Natl Acad Sci U S A. 2013;110:14278–14283. doi: 10.1073/pnas.1311845110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin ML, Julius JA, Tang X, Wang Y, Bachant J. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle. 2009;8:3406–3419. doi: 10.4161/cc.8.20.9911. [DOI] [PubMed] [Google Scholar]
- Bao J, Wang L, Lei J, Hu Y, Liu Y, Shen H, Yan W, Xu C. STK31(TDRD8) is dynamically regulated throughout mouse spermatogenesis and interacts with MIWI protein. Histochem Cell Biol. 2012;137:377–389. doi: 10.1007/s00418-011-0897-9. [DOI] [PubMed] [Google Scholar]
- Barysch SV, Dittner C, Flotho A, Becker J, Melchior F. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat Protoc. 2014;9:896–909. doi: 10.1038/nprot.2014.053. [DOI] [PubMed] [Google Scholar]
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomster HA, Imanishi SY, Siimes J, Kastu J, Morrice NA, Eriksson JE, Sistonen L. In vivo identification of sumoylation sites by a signature tag and cysteine-targeted affinity purification. J Biol Chem. 2010;285:19324–19329. doi: 10.1074/jbc.M110.106955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PW, Hwang K, Schlegel PN, Morris PL. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and SUMOylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum Reprod. 2008;23:2850–2857. doi: 10.1093/humrep/den300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M, Izumiya Y. Post-Translational Modifications of Kaposi's Sarcoma-Associated Herpesvirus Regulatory Proteins - SUMO and KSHV. Front Microbiol. 2012;3:31. doi: 10.3389/fmicb.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ding B, LeJeune D, Ruggiero C, Li S. Rpb1 sumoylation in response to UV radiation or transcriptional impairment in yeast. PLoS One. 2009;4:e5267. doi: 10.1371/journal.pone.0005267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupreta S, Holmstrom S, Subramanian L, Iniguez-Lluhi JA. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol Cell Biol. 2005;25:4272–4282. doi: 10.1128/MCB.25.10.4272-4282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P. Recent structural studies of RNA polymerases II and III. Biochem Soc Trans. 2006;34:1058–1061. doi: 10.1042/BST0341058. [DOI] [PubMed] [Google Scholar]
- Diril MK, Ratnacaram CK, Padmakumar VC, Du T, Wasser M, Coppola V, Tessarollo L, Kaldis P. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc Natl Acad Sci U S A. 2012;109:3826–3831. doi: 10.1073/pnas.1115201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok KL, Chung CM, Yi SQ, Jiang X, Sun X, Chen H, Chen YC, Kung HF, Tao Q, Diao R, Chan H, Zhang XH, Chung YW, Cai Z, Chang Chan H. STK31 maintains the undifferentiated state of colon cancer cells. Carcinogenesis. 2012;33:2044–2053. doi: 10.1093/carcin/bgs246. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- Hannoun Z, Greenhough S, Jaffray E, Hay RT, Hay DC. Post-translational modification by SUMO. Toxicology. 2010;278:288–293. doi: 10.1016/j.tox.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Herzog M, Wendling O, Guillou F, Chambon P, Mark M, Losson R, Cammas F. TIF1beta association with HP1 is essential for post-gastrulation development, but not for Sertoli cell functions during spermatogenesis. Dev Biol. 2011;350:548–558. doi: 10.1016/j.ydbio.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Hsiao HH, Meulmeester E, Frank BT, Melchior F, Urlaub H. “ChopNSpice,” a mass spectrometric approach that allows identification of endogenous small ubiquitin-like modifier-conjugated peptides. Mol Cell Proteomics. 2009;8:2664–2675. doi: 10.1074/mcp.M900087-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, Namekawa SH. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 2011;25:959–971. doi: 10.1101/gad.2030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O. SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 2007;8:550–555. doi: 10.1038/sj.embor.7400980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin M, Dmitrieva NI, Gallazzini M, Shen RF, Wang G, Burg MB, Ferraris JD. Mediator of DNA damage checkpoint 1 (MDC1) contributes to high NaCl-induced activation of the osmoprotective transcription factor TonEBP/OREBP. PLoS One. 2010;5:e12108. doi: 10.1371/journal.pone.0012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- La Salle S, Sun F, Handel MA. Isolation and short-term culture of mouse spermatocytes for analysis of meiosis. Methods Mol Biol. 2009;558:279–297. doi: 10.1007/978-1-60761-103-5_17. [DOI] [PubMed] [Google Scholar]
- La Salle S, Sun F, Zhang XD, Matunis MJ, Handel MA. Developmental control of sumoylation pathway proteins in mouse male germ cells. Dev Biol. 2008;321:227–237. doi: 10.1016/j.ydbio.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalmansingh AS, Urekar CJ, Reddi PP. TDP-43 is a transcriptional repressor: the testis-specific mouse acrv1 gene is a TDP-43 target in vivo. J Biol Chem. 2011;286:10970–10982. doi: 10.1074/jbc.M110.166587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko P. The DEAD-box helicase Vasa: evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim Biophys Acta. 2013;1829:810–816. doi: 10.1016/j.bbagrm.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, Ann DK. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282:36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- Li X, Lin HH, Chen H, Xu X, Shih HM, Ann DK. SUMOylation of the transcriptional co-repressor KAP1 is regulated by the serine and threonine phosphatase PP1. Sci Signal. 2010;3:ra32. doi: 10.1126/scisignal.2000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, Hung CC, Suen CS, Hwang MJ, Chang KS, Maul GG, Shih HM. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, Alt FW, Chen J. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Luo H, Zhou Y, Li Y, Li Q. Splice variants and promoter methylation status of the Bovine Vasa Homology (Bvh) gene may be involved in bull spermatogenesis. BMC Genet. 2013;14:58. doi: 10.1186/1471-2156-14-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R, Gerace L, Melchior F. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchiani S, Tamburrino L, Ricci B, Nosi D, Cambi M, Piomboni P, Belmonte G, Forti G, Muratori M, Baldi E. SUMO1 in human sperm: new targets, role in motility and morphology and relationship with DNA damage. Reproduction. 2014;148:453–467. doi: 10.1530/REP-14-0173. [DOI] [PubMed] [Google Scholar]
- Matafora V, D'Amato A, Mori S, Blasi F, Bachi A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol Cell Proteomics. 2009;8:2243–2255. doi: 10.1074/mcp.M900079-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler-Guillemain C, Depetris D, Luciani JJ, Mignon-Ravix C, Mitchell MJ, Mattei MG. In human pachytene spermatocytes, SUMO protein is restricted to the constitutive heterochromatin. Chromosome Res. 2008;16:761–782. doi: 10.1007/s10577-008-1225-7. [DOI] [PubMed] [Google Scholar]
- Moroianu J. Distinct nuclear import and export pathways mediated by members of the karyopherin beta family. J Cell Biochem. 1998;70:231–239. [PubMed] [Google Scholar]
- Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Myojin R, Kuwahara S, Yasaki T, Matsunaga T, Sakurai T, Kimura M, Uesugi S, Kurihara Y. Expression and functional significance of mouse paraspeckle protein 1 on spermatogenesis. Biol Reprod. 2004;71:926–932. doi: 10.1095/biolreprod.104.028159. [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nie M, Xie Y, Loo JA, Courey AJ. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS One. 2009;4:e5905. doi: 10.1371/journal.pone.0005905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Rogers RS, Inselman A, Handel MA, Matunis MJ. SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma. 2004;113:233–243. doi: 10.1007/s00412-004-0311-7. [DOI] [PubMed] [Google Scholar]
- Roscioli E, Di Francesco L, Bolognesi A, Giubettini M, Orlando S, Harel A, Schinina ME, Lavia P. Importin-beta negatively regulates multiple aspects of mitosis including RANGAP1 recruitment to kinetochores. J Cell Biol. 2012;196:435–450. doi: 10.1083/jcb.201109104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Detection of proteins sumoylated in vivo and in vitro. Methods Mol Biol. 2009;590:265–277. doi: 10.1007/978-1-60327-378-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava V, Marmor H, Chernyak S, Goldstein M, Feliciano M, Vigodner M. Cigarette smoke affects posttranslational modifications and inhibits capacitation-induced changes in human sperm proteins. Reprod Toxicol. 2014;43:125–129. doi: 10.1016/j.reprotox.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Shrivastava V, Pekar M, Grosser E, Im J, Vigodner M. SUMO proteins are involved in the stress response during spermatogenesis and are localized to DNA double-strand breaks in germ cells. Reproduction. 2010;139:999–1010. doi: 10.1530/REP-09-0492. [DOI] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhang Z, Hu W, Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J Biol Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- Stark GR, Taylor WR. Control of the G2/M transition. Mol Biotechnol. 2006;32:227–248. doi: 10.1385/MB:32:3:227. [DOI] [PubMed] [Google Scholar]
- Stopka T, Zakova D, Fuchs O, Kubrova O, Blafkova J, Jelinek J, Necas E, Zivny J. Chromatin remodeling gene SMARCA5 is dysregulated in primitive hematopoietic cells of acute leukemia. Leukemia. 2000;14:1247–1252. doi: 10.1038/sj.leu.2401807. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Ichiyama A, Saitoh H, Kawakami T, Omata M, Chung CH, Kimura M, Shimbara N, Tanaka K. A new 30-kDa ubiquitin-related SUMO-1 hydrolase from bovine brain. J Biol Chem. 1999;274:31131–31134. doi: 10.1074/jbc.274.44.31131. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- Tatham MH, Matic I, Mann M, Hay RT. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal. 2011;4:rs4. doi: 10.1126/scisignal.2001484. [DOI] [PubMed] [Google Scholar]
- Tatham MH, Rodriguez MS, Xirodimas DP, Hay RT. Detection of protein SUMOylation in vivo. Nat Protoc. 2009;4:1363–1371. doi: 10.1038/nprot.2009.128. [DOI] [PubMed] [Google Scholar]
- Vassileva MT, Matunis MJ. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol Cell Biol. 2004;24:3623–3632. doi: 10.1128/MCB.24.9.3623-3632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigodner M. Sumoylation precedes accumulation of phosphorylated H2AX on sex chromosomes during their meiotic inactivation. Chromosome Res. 2009;17:37–45. doi: 10.1007/s10577-008-9006-x. [DOI] [PubMed] [Google Scholar]
- Vigodner M, Ishikawa T, Schlegel PN, Morris PL. SUMO-1, human male germ cell development, and the androgen receptor in the testis of men with normal and abnormal spermatogenesis. Am J Physiol Endocrinol Metab. 2006;290:E1022–1033. doi: 10.1152/ajpendo.00527.2005. [DOI] [PubMed] [Google Scholar]
- Vigodner M, Morris PL. Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev Biol. 2005;282:480–492. doi: 10.1016/j.ydbio.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Vigodner M, Shrivastava V, Gutstein LE, Schneider J, Nieves E, Goldstein M, Feliciano M, Callaway M. Localization and identification of sumoylated proteins in human sperm: excessive sumoylation is a marker of defective spermatozoa. Hum Reprod. 2013;28:210–223. doi: 10.1093/humrep/des317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19:773–781. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Dixon SE, Ting LM, Liu TK, Jeffers V, Croken MM, Calloway M, Cannella D, Hakimi MA, Kim K, Sullivan WJ., Jr. Lysine acetyltransferase GCN5b interacts with AP2 factors and is required for Toxoplasma gondii proliferation. PLoS Pathog. 2014;10:e1003830. doi: 10.1371/journal.ppat.1003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gu H, Lin H, Chi T. Essential roles of the chromatin remodeling factor BRG1 in spermatogenesis in mice. Biol Reprod. 2012;86:186. doi: 10.1095/biolreprod.111.097097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dasso M. SUMOylation and deSUMOylation at a glance. J Cell Sci. 2009;122:4249–4252. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P, Cammas F, Gerard C, Metzger D, Chambon P, Losson R, Mark M. Germ cell expression of the transcriptional co-repressor TIF1beta is required for the maintenance of spermatogenesis in the mouse. Development. 2002;129:2329–2337. doi: 10.1242/dev.129.10.2329. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Pollack D, Nieves E, Winchell A, Callaway M, Vigodner M. Can your protein be sumoylated? A quick summary and important tips to study SUMO-modified proteins. Anal Biochem. 2014 doi: 10.1016/j.ab.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET. SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012;26:1196–1208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoe T, Tanaka F, Mimori K, Inoue H, Ohmachi T, Kusunoki M, Mori M. Efficient identification of a novel cancer/testis antigen for immunotherapy using three-step microarray analysis. Cancer Res. 2008;68:1074–1082. doi: 10.1158/0008-5472.CAN-07-0964. [DOI] [PubMed] [Google Scholar]
- Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xie M, Ren G, Yu B. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc Natl Acad Sci U S A. 2013;110:17588–17593. doi: 10.1073/pnas.1310644110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 2014;42:W325–330. doi: 10.1093/nar/gku383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. A histogram of the whole testis single-cell suspension prior to the STA-PUT procedure showed peaks of the haploid, diploid, and tetraploid cell populations (A). Representative images of fractions enriched for tetraploid and haploid cells after STA-PUT separation are shown. (B). Immunofluorescence experiments were carried out to examine the fraction purity and presence of sumoylated proteins upon completion of the prolonged separation procedure. SUMO was still localized to the centromeric heterochromatin (arrows) of spermatocytes and round spermatids, as well as to the sex body of spermatocytes (arrowheads). Scale bar is 10 μm.
Supplementary Figure 2. No significant difference was found between the results of a co-IP performed using denaturating and non-denaturating lysis buffers.