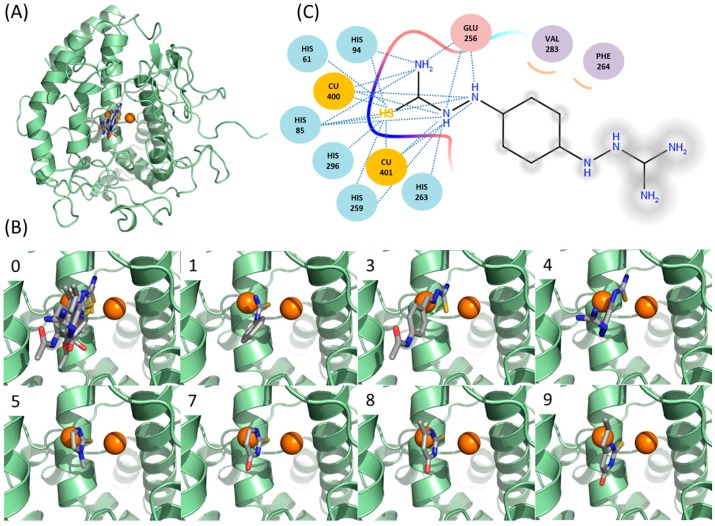
Figure 4.
Predicted binding modes of the thiourea-containing compounds. (A) Overlaid inhibitors against mushroom tyrosinase. Chain A of mushroom tyrosinase [15] (PDB: 2Y9X) are used for the docking with DOCK 3.6 [42]. The sphere and grids of the protein were prepared assuming the met-state of the copper atoms with the protocol by DOCK blaster [43]. The two copper atoms in tyrosinase are drawn as orange circles; (B) predicted binding mode in each inhibitory compound. (0) Overlay of the inhibitors in this study, (1) phenylthiourea, (3) thioacetazone, (4) ambazone, (5) methimazole, (7) thiourasil, (8) methylthiourasil, and (9) propylthiourasil. The yellow, red, and blue colors indicate the sulfur, oxygen, and nitrogen atoms, respectively. All the figures are prepared by Pymol [44] and arranged in the same orientation; and (C) schematic diagram of the representative interaction between ambazone and tyrosinase. Residues in tyrosinase are drawn in circles. Residues for hydrophilic and hydrophobic interactions are coloured in pink and purple, respectively. Dashed lines correspond to hydrophilic interactions. Curves indicate hydrophobic contacts. Atoms exposed to solvent exposure are shaded.