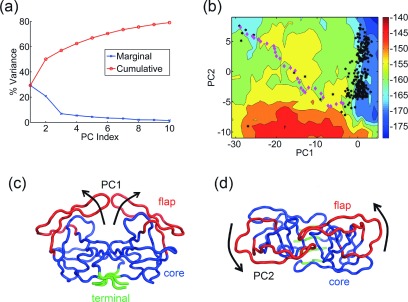
FIG. 5.
Predicted conformational transition pathway for HIV-1 protease. (a) Percentage of variance captured by the first 10 PCs from the set of 304 HIV-1 protease structures. (b) Visualization of PC1—opening and closing of the flap domains (red) against the core domain (blue). The terminal domain is shown in green. (c) Visualization of PC2—twisting motion of the flaps (red). (d) Free energy landscape of the molecule along PC1-PC2 (entropy weight a = 1.3). Crystal structures are denoted by black hexagons, while intermediate structures along the predicted transition pathway are shown as magenta diamonds. The predicted transition pathway follows a relatively low-energy path on the landscape along a diagonal path and passes close to several experimental intermediate forms.