Abstract
In this work, we present the first XOS degrading glycoside hydrolase from Weissella, WXyn43, a two-domain enzyme from GH43. The gene was amplified from genomic DNA of the XOS utilizing Weissella strain 92, classified under the species-pair Weissella cibaria/W.confusa, and expressed in Escherichia coli. The enzyme is lacking a putative signal peptide and is, from a homology model, shown to be composed of an N-terminal 5-fold β-propeller catalytic domain and a C-terminal β-sandwich domain of unknown function. WXyn43 hydrolyzed short (1–4)-β-d-xylooligosaccharides, with similar kcat/KM for xylobiose (X2) and xylotriose (X3) and clearly lower efficiency in xylotetraose (X4) conversion. WXyn43 displays the highest reported kcat for conversion of X3 (900 s−1 at 37°C) and X4 (770 s−1), and kcat for hydrolysis of X2 (907 s−1) is comparable with or greater than the highest previously reported. The purified enzyme adopted a homotetrameric state in solution, while a truncated form with isolated N-terminal catalytic domain adopted a mixture of oligomeric states and lacked detectable activity. The homology model shows that residues from both domains are involved in monomer–monomer hydrogen bonds, while the bonds creating dimer–dimer interactions only involved residues from the N-terminal domain. Docking of X2 and X3 in the active site shows interactions corresponding to subsites −1 and +1, while presence of a third subsite is unclear, but interactions between a loop and the reducing-end xylose of X3 may be present.
Keywords: prebiotics, structure–function, W. cibaria, W. confusa, xylosidase
Introduction
A balanced composition of the gut microbiota plays an important role for health and prevention of disease, a topic that has been extensively reviewed by several authors (Sekirov et al. 2010; Wallace et al. 2011; Clemente et al. 2012). To obtain this balance, health promoting bacteria, probiotics, are important for modulating microbial composition to protect against infections and stimulate immune function (Ohland and MacNaughton 2010). Most bacterial strains classified as probiotics thus far belong to either bifidobacteria or lactobacilli (Kleerebezem and Vaughan 2009), but other genera of lactic acid bacteria (LAB) commonly found in fermented-foods and in the gastrointestinal tract of humans also contain probiotic candidates. One example is strains from the species-pair Weissella cibaria/confusa, isolated from food products. Candidates from these species have shown good resistance to acid and salts as well as good adhesion to human colon Caco-2 cells, supporting probiotic characteristics (Lee et al. 2012). Recently, we have also shown that some strains, including the currently investigated strain 92, are able to utilize xylooligosaccharides (XOS) derived from hardwood (Patel et al. 2013) as well as un-substituted XOS derived from wheat bran (Immerzeel et al. 2014).
XOS are one of the most promising types of emerging prebiotics. Prebiotics are nondigestible oligosaccharides that selectively stimulate growth and/or metabolic activity of probiotic bacteria (Crittenden et al. 2002; Roberfroid 2007; Gullon et al. 2008; Rastall 2010; Falck et al. 2013). Fermentation of prebiotics by probiotics leads to the formation of short chain fatty acids, which can reduce growth of pathogens, and in the case of butyrate also protect against colon cancer (Hijova and Chmelarova 2007). XOS are composed of (1 → 4)-β-linked d-xylopyranosyl units with a degree of polymerization in the range 2–10. Substituted XOS can be derived from cereals, with (1 → 2) and/or (1 → 3)-α-linked l-arabinofuranosyl (Araf) substituents [arabinoxylooligosaccharides (AXOS)]. In addition, AXOS can carry acetyl groups, α-d-glucopyranosyluronic acid, its 4-O-methyl derivative and hydroxycinnamic acid derivatives (Moure et al. 2006; Aachary and Prapulla 2011; Broekaert et al. 2011).
Degradation of hemicelluloses with differently substituted xylans depends on activities from a number of different glycoside hydrolases (GHs). These GHs are classified as xylanases (GH5, GH10, GH11), β-xylosidases (GH3, GH39, GH43), exo-oligoxylanases (GH8), arabinofuranosidases (GH43, GH51), α-glucuronidases (GH67, GH115), as well as a number of esterases, acetyl xylan esterases (CE1, CE2, CE4, CE6, CE7) and feruloyl esterases (CE1) (Lagaert et al. 2007; Flint et al. 2012). The XOS degrading β-xylosidases can be classified as either retaining (GH3, GH39) or inverting (GH43) depending on whether the catalytic mechanism involves a two-step double (retaining) or a one-step single displacement reaction (inverting), respectively. Degradation of AXOS requires additional activities from α-l-arabinofuranosidases (Arafases) (EC 3.2.1.55) able to release Araf substituents linked to the main chain. Depending on the linkage that is cleaved between the xylose and arabinose, as well as activity on either single or double substituted xylose units, different activity profiles can be distinguished for Arafases (Lagaert et al. 2010).
In a previous work, we have shown that the genes encoding putative xylan degrading enzymes in available genome sequences of W. cibaria and W. confusa have similarities with GH43 (Patel et al. 2013), but none of these GHs have to date been characterized from either of these two species. Among probiotics, bifidobacteria have been the most studied genus with respect to their β-xylosidases (van den Broek and Voragen 2008; Lagaert et al. 2011; Amaretti et al. 2013). Enzyme candidates from genera under the order Lactobacillales have been less investigated and particularly we completely lack information on GHs from the genus Weissella. Recently, two GH43 β-xylosidases from Lactobacillus brevis have been characterized (Michlmayr et al. 2013), shedding some light on these enzymes in a related genus classified under Lactobacillales. Enzymes from GH43 have Arafase or β-d-xylosidase activity or both activities (bifunctional), often displaying both activities on aryl-glycoside substrates.
The first crystal structure for family 43 was reported for CjAbn43A from Cellvibrio japonicus (Nurizzo et al. 2002). This protein displayed a novel five-bladed-β-propeller structure, where the propeller blades are composed of four β-sheets, and binding to the substrate occurs in a long V-shaped groove located across the surface of the propeller. The catalytic general base is an aspartate and the general acid a glutamic acid. The variation in substrate specificity observed in GH43 has been explained to depend on the different ways the substrate orients in the deep pocket.
In this work, we present cloning, production, and characterization of WXyn43, a new GH43 from the XOS utilizing and putative probiotic Weissella strain 92. We have investigated the oligomeric state of the enzyme, along with its substrate specificity, optimal temperature, and pH range and kinetic parameters. A homology model was also created based on crystallographic structures of β-xylosidases from Selenomonas ruminantium (PDB 3C2U), Geobacillus stearothermophilus (PED 2EXI), and Bacillus halodurans (PDB 1YRZ). Based on its high-sequence homology to other known GH43 enzymes and its activity on XOS, WXyn43 is classified as a GH43 β-d-xylosidase. New insight into the function and structure of these GH43 could also widen the knowledge about the β-xylosidases in LAB in general.
Results
Expression, sequence analysis and oligomeric state
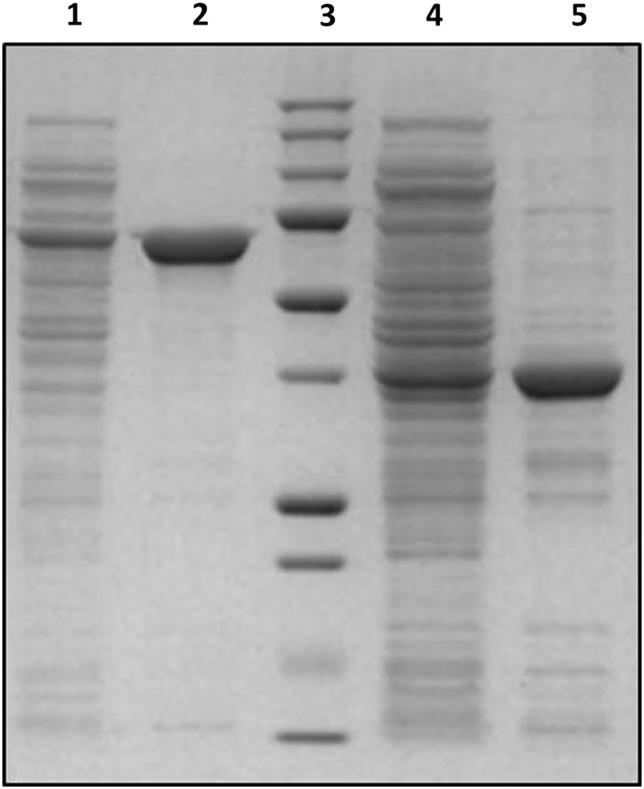
A gene (GenBank: KP903368) encoding a β-xylosidase from Weissella sp. strain 92 was cloned and the corresponding recombinant-protein was produced in E. coli BL21(DE3). Expression of the gene encoding WXyn43, and a truncated gene encoding the catalytic domain (347 amino acids with a theoretically determined molecular mass of 39107.3 Da), resulted in high-level productions of the respective protein, corresponding to >50% of the total protein (Figure 1).
Fig. 1.
SDS–PAGE of expressed and purified proteins. Lane 1: crude WXyn43, lane 2: WXyn43, lane 3: molecular marker (from the top: 250, 150, 100, 75, 50, 37, 25, 20, 15 and 10 kDa), lane 4: crude WXyn43 truncated catalytic domain and lane 5: WXyn43 truncated catalytic domain.
The deduced amino acid sequence of the full-length enzyme consists of 583 residues (including the N-terminal histidine tag) corresponding to a molecular mass of 65978.6 Da, which is in accordance with the apparent molecular mass estimated by SDS–PAGE (Figure 1). Analysis by SignalP (www.cbs.dtu.dk/services/SignalP/) showed the absence of any likely signal peptide, suggesting that the enzyme is intracellular.
Blastp analysis of the amino acid sequence revealed 99% identity to a putative GH43 xylosidase encoded by a gene reported from a genome-sequencing project of W. confusa LBAE C39-2 (Amari et al. 2012) (NCBI Reference Sequence: WP_003610085.1). No GHs from the genus Weissella are however characterized to date, and the gene (identified from an ongoing sequencing project) is not yet registered in the carbohydrate active enzyme database (www.cazy.org). According to the Blastp report, the sequence conservation in the N-terminal part is typical for catalytic domains of GH43 β-xylosidases (EC 3.2.1.37). This part is followed by an additional domain in the C-terminus. Alignment of the sequence from Weissella strain 92 with that of the putative xylosidase fromW. confusa LBAE C39-2 showed that only three residues differ: one in the N-terminal catalytic domain (Asp274 in strain 92, corresponding to Glu in LBAE C39-2) and two (Asp400 and Asp429 in strain 92, corresponding to Asn400 and Glu429 in LBAE C39-2) in the part corresponding to the C-terminal domain. (Modelling of the structure, below, showed that none of the three residues was located in the predicted active-site cavity, suggesting that they do not affect substrate specificity.) Sequence identity to enzymes of known function and 3D-structure was 53% to the GH43 of S. ruminantium, 47% to a GH43 enzyme from G. stearothermophilus and 47% to a GH43 from B. halodurans, allowing unambiguous classification under GH43. The level of identical residues allowed modelling of the 3D-structure, and more detailed identification of domain boundaries and residues important for activity (see sections on Modelled overall structure and Prediction of subsites). Based on the above results, the enzyme from Weissella sp. strain 92 is hereafter named WXyn43.
Molecular mass determination of WXyn43 by size exclusion chromatography (SEC) gave a single peak estimated to a molecular mass of 259 kDa (Supplementary data, Fig. S1 and Table SIV), consistent with a homo-tetramer complex. This differed from the SEC profile of the truncated form (encoding the catalytic domain only), which showed four peaks (Supplementary data, Table SIV), suggesting the presence of monomers as well as complexes of dimers or trimers and tetramers, indicating weaker interactions between the subunits of the truncated form.
Activity studies
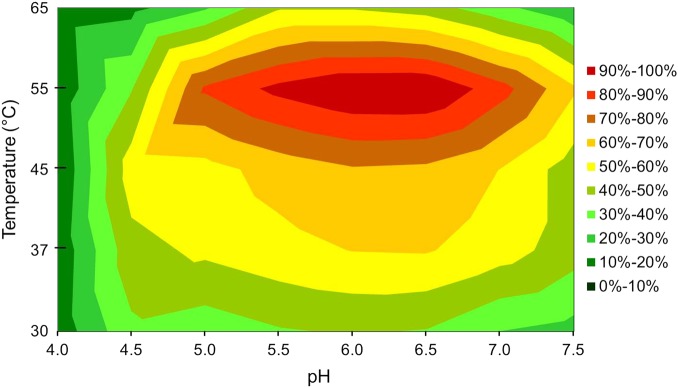
Measurements of the β-xylosidase activity on p-nitrophenyl-β-d-xylopyranoside (pNPX) over a range of different temperatures and pHs in a surface plot showed that WXyn43 was active in the broad pH range 5–7, with an optimum between 6.0 and 6.5 (Figure 2). Highest activity was determined at 55°C. However, at 55°C the stability of the enzyme was very poor, as seen by the half-life of 4 min. At 45°C the enzyme lost 40% activity after 60 min, while at 37°C the enzyme was completely stable for at least 60 min. Due to the high stability at 37°C, which is also the growth temperature of the Weissella strain (and the temperature in the gut), this temperature was chosen for activity and kinetic measurements. Specific activity measurements on pNPX, p-nitrophenyl-α-l-arabinofuranoside (pNPAf) and oligosaccharide substrates showed that WXyn43 was active on pNPX, pNPAf and on XOS with a degree of polymerization (DP) 2–4 (Table I). Activity was 20-fold higher on pNPX (11.2 U/mg) than on pNPAf (0.51 U/mg), which shows that the expressed protein is a true β-xylosidase, with a minor pNPAf side-activity. No Arafase activity was detected for the natural substrates, namely (1 → 5) linked arabinooligosaccharides (AOS) and (1 → 2) or (1 → 3) arabinose substituted AXOS. Neither xylose nor arabinose was released from AXOS at the specific reaction conditions used in the assay. Activity on (1 → 4)-β-linked XOS decreased drastically for xylotetraose (X4) compared with xylobiose (X2) and xylotriose (X3) indicating that the size of the substrate plays an important role for the binding and accessibility to the active site (Table I). No pNPX activity was detected using the truncated form of the enzyme (data not shown), showing that the second β-sandwich domain is important for catalytic activity.
Fig. 2.
Surface of WXyn43 activity on p-nitrophenyl-β-d-xylopyranoside at different pH and temperatures. This figure is available in black and white in print and in color at Glycobiology online.
Table I.
WXyn43-specific activity for 1 mM p-nitrophenyl and oligosaccharide substrates
| Substrate | Specific activity (U/mg) |
|---|---|
| p-Nitrophenyl-α-l-arabinofuranoside | 0.51 ± 0.01 |
| p-nitrophenyl-α-d-galactopyranoside | n.d. |
| p-nitrophenyl-β-d-glucopyranoside | n.d. |
| p-Nitrophenyl-β-d-mannopyranoside | n.d. |
| p-Nitrophenyl-β-d-xylopyranoside | 11.2 ± 0.7 |
| (1 → 4)-β-d-Xylobiose | 100 ± 5 |
| (1 → 4)-β-d-Xylotriose | 98 ± 4 |
| (1 → 4)-β-d-xylotetraose | 29 ± 2 |
| (1 → 5)-α-l-Arabinobiose | n.d. |
| (1 → 5)-α-l-Arabinotriose | n.d. |
| (1 → 3)-l-Arabinosyl-β-d-xylobiose | n.d. |
| (1 → 2)-Arabinosyl-β-d-xylotriose | n.d. |
n.d., not detected; U, µmol/min.
Kinetics
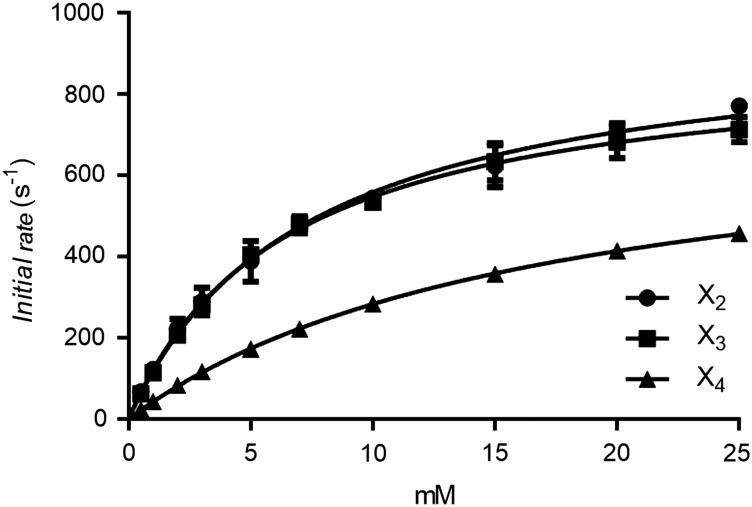
The kinetics was measured as the initial rate of product formation from synthetic and natural substrates. This resulted in data that could be fitted by nonlinear regression to the Michaelis–Menten equation (Table II). The turnover and catalytic efficiency for X2 and X3 were very similar, while both were significantly lower for X4 (Table II and Figure 3). The kcat/KM shows that WXyn43 is three times as efficient in conversion of the short substrates X2 and X3 compared with X4. Comparison of KM and kcat values with those of enzymes from the related bacterium L. brevis (Weissella and Lactobacillus are both classified under the order Lactobacillales, but in different families) previously reported in the literature, including XynB2 (at 37°C) from L. brevis DSM 20054 (Michlmayr et al. 2013) and Lbx (at 25°C) from L. brevis ATCC 367 (Jordan et al. 2013), shows that WXyn43 has a higher turnover (kcat), which is especially striking using X3, and which may compensate for the higher KM observed (Table II).
Table II.
WXyn43 kinetic constants in comparison with reported data on β-xylosidases from XynB2 (Michlmayr et al. 2013) and LbX (Jordan et al. 2013), both from strains of L. brevis
| β-Xylosidase | Substrate | Kinetic constant |
||
|---|---|---|---|---|
| kcat (s−1) | KM (mM) | kcat/KM (s−1 mM−1) | ||
| WXyn43 | ||||
| p-Nitrophenyl-β-d-xylopyranoside | 258 ± 11 | 7.4 ± 1.1 | 34.9 ± 5.4 | |
| (1 → 4)-β-d-Xylobiose | 961 ± 25 | 7.2 ± 0.5 | 133.5 ± 9.9 | |
| (1 → 4)-β-d-Xylotriose | 900 ± 13 | 6.5 ± 0.3 | 138.5 ± 6.7 | |
| (1 → 4)-β-d-Xylotetraose | 770 ± 7 | 17 ± 0.3 | 45.3 ± 0.9 | |
| XynB2a | ||||
| p-Nitrophenyl-β-d-xylopyranoside | 46 | 11 ± 1 | 4.1 | |
| (1 → 4)-β-d-Xylobiose | 233 | 4.8 ± 0.4 | 48 | |
| LbXb | ||||
| p-Nitrophenyl-β-d-xylopyranoside | 73.4 ± 1.5 | 6.93 ± 0.26 | 10.6 ± 0.2 | |
| (1 → 4)-β-d-Xylobiose | 407 ± 9 | 2.96 ± 0.24 | 138 ± 9 | |
| (1 → 4)-β-d-Xylotriose | 235 ± 4 | 2.91 ± 0.15 | 80.8 ± 3.0 | |
| (1 → 4)-β-d-Xylotetraose | 146 ± 4 | 2.40 ± 0.08 | 32.6 ± 1.2 | |
aReactions performed at 37°C and pH 5.5 and b25°C and pH 6.0.
Fig. 3.
WXyn43 nonlinear regressions to the Michaelis–Menten equation at 37°C and pH 6.0. Error bars represent the standard deviation for n = 3. X2: xylobiose, X3: xylotriose and X4: xylotetraose.
Modelled overall structure and predicted catalytic residues
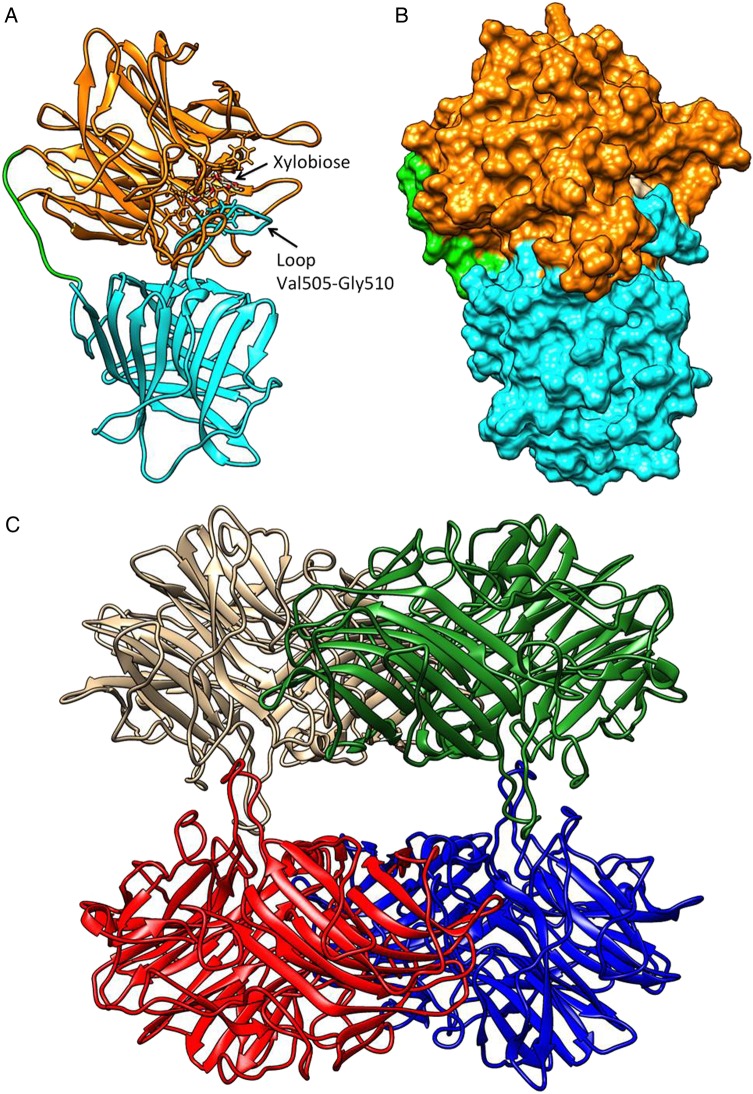
Homology hybrid models of a single subunit (protomer, Figure 4A and B) and a homo-tetramer (Figure 4C) were built for WXyn43, confirming the two-domain structure composed of an N-terminal five-bladed (I to V) β-propeller catalytic domain (Met1-Lys320) connected to a C-terminal β-jelly roll-folded domain (His331–Gly543) via a long loop (Asp321–Asp330).
Fig. 4.
WXyn43 homology models for monomers of the two-domain enzyme (A and B), and the tetrameric form (C). In (A) the catalytic domain is shown in orange, the C-terminal domain in cyan and the inter-domains loop in green. A xylobiose is bound in the active site, and the loop putatively interacting with xylotriose is indicated. In (B), the active-site cavity is shown in surface representation using the same color scheme. In (C), the hybrid homology model of the homo-tetramer is shown: chain A in brown, chain B in blue, chain D in green and chain C in red. Interactions between chains A–D and B–C make compact dimers, while the interactions A–C and B–D stabilize the tetramer (see also Supplementary data, Table SV). Molecular graphics and analyses were performed with the UCSF Chimera v1.8.1 package (Pettersen et al. 2004).
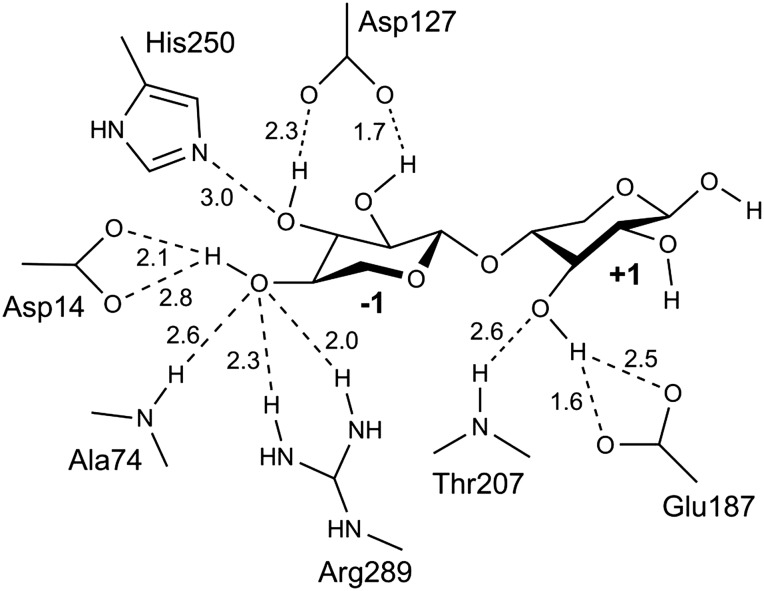
The active site is located in the center of the five-bladed β-propeller, and the catalytic residues are conserved for enzymes classified under Clan-F (GH43 and GH62) and the related Clan-J (GH32 and GH68) (Nurizzo et al. 2002; Brüx et al. 2006). GH43 is a family of inverting enzymes, and the general acid is predicted to be Glu187 (situated 3.0 Å from the glycosidic oxygen) while Asp14 is predicted as the general base (situated 6.0 Å from the anomeric carbon). A catalytic residue modulating the pKa of the general acid and its orientation in relation to the substrate, is essential for most five-bladed β-propeller GHs (Nurizzo et al. 2002; Brüx et al. 2006). In the model of WXyn43, Asp127 (located 5.5 Å from Glu187) is proposed for this role (Figure 5).
Fig. 5.
Active site in the homology model of WXyn43. Schematic representation of potential hydrogen bonds, the distances are given in angstrom.
The exact function of the C-terminal β-jelly roll-folded domain is not known, but alignment studies using the β-xylosidase XynB3 from G. stearothermophilus describe that a conserved Phe-residue in the C-domain (Phe513 in WXyn43) is part of the substrate-interacting residues in the active site (Brüx et al. 2006), which may be part of the explanation for the lack of detectable pNPX activity using the truncated protein. Moreover, the C-terminal domain has a role in interprotomeric interactions (Figure 4C). The homo-tetramer can be described as a dimer of dimers. This means two protomers tightly arranged as a compact dimer (via monomer–monomer interactions), which in turn interacts with another compact dimer (via dimer–dimer interactions) through long loops leaving a central hole that allows the substrate to enter into the active-site pocket of each subunit. Based on the model, monomer–monomer interactions involve hydrogen bonds between conserved residues in loops from both the β-propeller domain and the β-jelly roll-folded domain of the neighboring subunit (Supplementary data, Table SV) while dimer–dimer interactions (stabilizing the tetramer) involve less conserved residues in three loops from the β-propeller domain.
Prediction of subsites
Two substrate binding subsites (−1 and +1) were identified after modeling X2 into the active-site pocket. X2 did not undergo significant conformational changes upon modelling into the active site, and the interactions with the amino acids in the active site were similar to those observed in the crystal complex of XynB3 (PDB, 2EXJ).
Subsite −1 determines glycone specificity [in GH43 for either xylose, arabinose, or galactose (www.cazy.org/GH43.html)], and in WXyn43 mainly involves interactions with xylose hydroxyl groups via hydrogen bonding to residues Asp14, Ala74, Asp127, His250 and Arg289 (Figure 5). Hydrophobic and stacking substrate interactions in this subsite are predicted to involve residues Phe26, Phe31, Trp73 and Phe513.
The aglycone xylose is less tightly bound in subsite +1. Only the O3-hydroxyl of the aglycone xylose is potentially bound via hydrogen bonds with Glu187 and Thr207, while hydrophobic interactions could involve Phe154, Phe512 and Phe513.
The presence of additional +2 subsite interactions (explaining the more beneficial turnover of xylotriose in WXyn43 compared with the L. brevis enzymes) is unclear, but may in WXyn43 include interaction with a loop (Val505–Gly510).
Discussion
XOS are getting increased attention as putative prebiotics, important for sustaining and increasing the number of beneficial genera in the gastrointestinal tract in order to improve health and prevent disease. Certain species of LAB have the ability to utilize XOS but not xylan as carbon source (Crittenden et al. 2002; Flint et al. 2012; Patel et al. 2013), and XOS can subsequently be used to promote their growth. To increase the understanding of how the XOS are metabolized it is of interest to map and characterize the enzymes responsible for XOS degradation in putative probiotic strains, and in this work we have characterized a first enzyme from the putative probiotic strain 92, from the species Weissella. Inspection of the distribution of XOS-degrading enzymes in the CaZy database (www.cazy.org) shows that such enzymes can be found classified as xylanases (GH5, GH10, GH11), β-xylosidases (GH3, GH43), or exo-oligoxylanases (GH8). We have previously found that genome sequences of W. cibaria (strain KACC 11862) and W. confusa (strain LBAE C39-2) encode genes with similarities to GH43, while no genes encoding putative xylanases from GH5, 10 or 11 can be found, and genes encoding putative GH3 and 8 are unevenly distributed (Patel et al. 2013).
In this work, we have shown that WXyn43, a GH43 β-xylosidase from the newly isolated Weissella sp. strain 92 (Patel et al. 2013), by 16S sequencing classified under the species-pair W. cibaria/confusa), is indeed an exoacting XOS converting enzyme. Activity studies showed that the enzyme is active on both pNPX and pNPAf (Table I), while studies on natural arabinofuranosidase substrates (arabinose 1 → 2 or 1 → 3 linked to xylose in AXOS or with 1 → 5 linkages in AOS) revealed that the enzyme was lacking detectable activity towards these substrates. In the AXOS, arabinose residues were attached to the nonreducing-end xylose, and neither xylose nor arabinose was released from these substrates (GH43 β-xylosidases, EC-number 3.2.1.37, act from the nonreducing-end releasing single xylose units) showing that the arabinose side group effectively hinders binding of the substituted xylose in the −1 subsite, and that the 1 → 2- or 1 → 3-linked Araf substituents cannot take the place of the xylose in the subsite.
Kinetic studies on non-substituted xylose substrates (both pNPX and natural XOS) showed that WXyn43 has a higher kcat but a lower substrate affinity (KM) at 37°C compared with either XynB2 at 37°C (Michlmayr et al. 2013) or LbX at 25°C (Jordan et al. 2013). WXyn43 displays the highest reported kcat values for X3 and X4 hydrolysis, and kcat (X2) (961 s−1 at 37°C) is comparable with or greater than the highest previously reported for LbX (407 s−1 at 25°C; Table II). It seems that the weaker affinity is compensated by a higher turnover number, which might also result in lower product inhibition. Specific activity and kinetic measurements for X2 and X3 were very similar under the reaction conditions tested, and only for X4 was the activity significantly lower (Tables I and II). This is different to what has been reported for the two β-xylosidases from L. brevis strains (Jordan et al. 2013; Michlmayr et al. 2013), which displayed higher activity on X2 compared with X3, indicating that these enzymes only have two binding sites. In the case of WXyn43 the activity against X3 could potentially be improved by interactions with the loop (Val505–Gly510 from the C-terminal domain) at the reducing end of X3. These potential interactions could explain the increase in turnover for X3, which is very similar to the one of X2. The reduced activity on X4 indicate that this substrate has no additional interactions with the enzyme, and that instead this substrate and substrates of higher DP could be sterically restrained based on surrounding structures in the tetrameric complex. The results obtained from XOS kinetics also correlate well to the XOS utilization pattern observed in fermentation studies of strain 92, where depletion of X2 and X3 was significantly faster than for X4 (Patel et al. 2013). Thus the evolutionary pressure to utilize substrates longer than X3 may be low as the uptake of longer XOS is slow.
Crystallographic structures of β-xylosidases in family GH43 (www.cazy.org) show enzymes with two general domain-compositions: two-and single-domain enzymes. Two-domain enzymes of known structure are shown in Table III, including WXyn43 modeled in the present work. All these structures are tetramers, except the B. halodurans C-125 (PDB 1YRZ) β-xylosidase, which is a dimer. The role of the oligomerization is not yet clear. Only two structures were co-crystallized with ligand, β-xylosidase from G. stearothermophilus (PDB 2EXJ) with the natural substrate β-d-xylobiose (Brüx et al. 2006) and β-xylosidase from S. ruminantium in complex with the inhibitor 1,3-bis[tris(hydroxymethyl) methylamino]propane (PDB 3C2U) (Brunzelle et al. 2008). However, all structurally characterized two-domain enzymes conserve the residues potentially involved in hydrogen bond interactions in both subsites such as represented in the modelled interaction shown in Figure 5. Thus, these structural features together with the kinetic analysis, and the intracellular location (none are reported to contain a signal peptide) show that they are specialized in the hydrolysis of short substrates, taken up by the respective microorganism.
Table III.
Comparison of the β-xylosidase modeled (WXyn43) with the crystallographic structures of two-domain β-xylosidases from family GH43
| Source | PDB code | Identities (%) | Positives (%) | Gaps (%) | Residues aligneda | Overall RMSD (Å)a |
|---|---|---|---|---|---|---|
| Weissella sp. strain. 92 | Model | 100 | 100 | 0 | 553 | 0 |
| Selenomonas ruminantium | 3C2U | 53 | 65 | 3 | 526 | 0.936 |
| Geobacillus stearothermophilus | 2EXI | 47 | 64 | 2 | 526 | 0.896 |
| Bacillus halodurans | 1YRZ | 47 | 61 | 5 | 513 | 1.137 |
| Bacillus subtilis subsp. subtilis | 1YIF | 49 | 64 | 3 | 488 | 0.788 |
| Clostridium acetobutylicum | 1Y7B | 51 | 67 | 2 | 501 | 0.784 |
3C2U, 2EXI and 1YRZ were used as templates for the hybrid model. Sequence similarities to WXyn43, and Cα-RMSD of the superimposed structures are given.
aThe overall RMSD was calculated from the residue aligned after superimposing the structures with a match alignment cutoff of 5.0 Å.
The single-domain GH43 enzymes consist only of the five-bladed β-propeller domain. Structure determined xylosidases of this type have been obtained from Clostridium acetobutylicum ATCC 824 (PDB 3K1U) and from an uncultured sample (PDB 4MLG). In addition, one single-domain enzyme with endo-β-1,4-xylanase activity from Bacteroides thetaiotaomicron VPI-5482 has had its structure determined (PDB 3QZ4). No kinetic data are however published for these enzymes, making comparisons of the influence of the C-domain on the activity profile difficult. However, comparison of substrate-interacting residues in the single- and two-domain enzymes shows that only the catalytic residues (Asp, Glu, and its pKa-modulating Asp) are well conserved, suggesting that the substrates could bind in different manners. In addition, a potential signal peptide has been reported for the endo-β-1,4-xylanase encoding gene from Bacteroides thetaiotaomicron VPI-5482, suggesting that this is an extracellular enzyme, which is consistent with degradation of long substrates. The truncated Weissella β-xylosidase, encoding the catalytic domain is predicted to share the same fold as the structures mentioned above; however, no activity was detected using this construct. According to the predictions, the truncated form shares most of the potential residues involved in substrate binding, but has lost the Phe513 residue of the C-domain, indicating that this residue may be fundamental for binding of xylobiose in subsite −1, and that naturally occurring single-domain enzymes has solved this by using a different set-up of substrate-interacting residues. In addition, full-length WXyn43 has only one form in solution, the homo-tetramer complex, while the truncated catalytic domain has multiple forms, showing that the C-terminal domain is important for oligomerization of this enzyme. The role of oligomerization in two-domain GH43 enzymes is however still unclear. As a comparison, XynB2 from L. brevis DSM 20054 (Michlmayr et al. 2013) is reported as a dimer but has similar properties to WXyn43. The interaction analysis indicated that both the catalytic and the C-terminal domain of the monomer take part in the monomer–monomer to dimer interactions, while only the catalytic domain is involved in the dimer–dimer to tetramer interaction. This combined with the results on the active L. brevis dimer indicate that the monomer–monomer interactions (also involving substrate interactions) may be most crucial for activity.
Similarity searches by Blastp, using the amino acid sequence of WXyn43 as query sequence, showed that sequences similar to this β-xylosidase are not restricted to putative probiotic bacteria, but are occurring in probiotics as well as in some human and animal pathogens. The potential probiotic organisms harboring related putative GH43 β-xylosidase genes were, based on the bioinformatics search, species of Leuconostoc, Lactobacillus, Lactococcus, and Bifidobacterium, while the potential pathogens were Mannhemia haemolytica, Klebsiella oxytoca, Klebsiella pneumoniae and Enterobacter aerogenes. While the probiotics are Gram positive, the pathogens are Gram negative, and the distribution of this GH family in both groups could suggest horizontal gene transfer. However, XOS have only been reported to stimulate the growth of nonpathogenic bacteria indicating a difference in utilization of these substrates by the probiotics and potentially harmful bacteria that may be related to uptake of the substrate. Homologous proteins for WXyn43 were found in Leuconostoc, Lactococcus and Lactobacillus using Blastp. This shows that the knowledge obtained in this study could be applied to elucidate XOS utilization in related genera.
Conclusions
In this work, the first GH43 β-xylosidase (WXyn43) from Weissella was characterized. The enzyme hydrolyzes X2 and X3 with the same efficiency, indicating possible interactions with a flexible loop at the reducing end of X3. Hydrolysis of X4 is significantly reduced possibly due to unfavorable interactions with the substrate. The change in kcat/KM with DP of XOS substrate for WXyn43 matches the carbohydrate preference of strain 92 in previously performed fermentation experiments. The C-terminal β-sandwich domain is shown to be important both for oligomerization and activity, indicating that at least a dimeric form may be necessary for activity. More research is however needed to conclude the role of the C-terminal domain in the oligomerization and for the activity of this and related β-xylosidases and whether the presence of the C-terminal domain has implications on the length of the substrates utilized by the enzyme.
Materials and methods
Substrates and reaction buffers
All p-nitrophenyl-glycoside substrates were obtained from Carbosynth (Berkshire, UK). XOS and AXOS were all from Megazyme (Wicklow, Ireland). AXOS standards (1 → 3)-arabinosyl-xylobiose and (1 → 2)-arabinosyl-xylotriose were kindly supplied by Megazyme. Both AXOS standards had the arabinose unit attached to the terminal nonreducing-end xylose.
Strains, plasmids and culture medium
The Weissella sp. strain 92 was obtained from our species collection (Patel et al. 2013) and was by 16S rDNA sequencing shown to be a member of the species-pair W. cibaria/confusa [indistinguishable by 16S sequencing (Patel et al. 2013)]. Escherichia coli strain NovaBlue from Novagen was used as cloning host and strain BL21(DE3) was used for expression. Cultivation of E. coli was performed in Luria Bertani (LB) broth and on agar plates at 37°C. Propagation vector pUC19 (Thermo scientific, Gothenburg, Sweden) was used for cloning, and the recombinant clones were selected using LB ampicillin (100 µg/mL). The gene was subcloned into the expression vector pET28b (Novagen, affiliate of Merck, Darmstadt, Germany) and the clones were selected from LB kanamycin (30 µg/mL) plates. Antibiotics were purchased from Sigma-Aldrich (Steinheim, Germany).
Cloning
All enzymes used in the cloning were purchased from Thermo Scientific and all kits were purchased from Qiagen (Hamburg, Germany). Genomic DNA from Weissella sp. strain 92 was extracted as described by Patel et al. (2012) and used as a template to amplify a putative β-xylosidase. Primers were designed based on a putative xylosidase encoded in the genome of W. confusa (Amari et al. 2012) (5′–3′), Fw: CATATGATGCAAATTCAAAACCCTGTATTGCC and Rv: CTCGAGTTAGCCGAAGCGTGATTCTGAAGC. The PCR products were cleaned up using the FAST version of AP and ExoI and the PCR fragments were ligated in the vector pUC19 for its propagation in E. coli NovaBlue. The clone selection was done on LB plates containing 100 µg/mL ampicillin and screened for positive clones by the α-complementation technique using X-gal (blue/white colonies). Insertion of the target gene was confirmed by colony PCR. The positive plasmids were digested using NdeI and XhoI, the fragments were extracted from an agarose using an extraction kit. The fragments were then ligated with the expression vector giving the final construct pET28b-WXyn43 for expression of the gene encoding the native protein with an N-terminal LE-6 × His-sequence. (The two additional amino acids (Leu, Glu) are a result of the XhoI-site.) The expression vector was transformed into E. coli BL21(DE3) competent cells. Clones were picked based on colony PCR to confirm the insert. The positive clones were finally confirmed by DNA sequencing (GATC Biotech, Konstanz, Germany). The sequence of the obtained gene is deposited in GenBank under the accession number KP903368.
Mutational deletion of the β-sandwich domain
The catalytic domain was obtained by amplification from the final construct (pET28b-WXyn43) removing the N-terminal domain using primers (5′–3′), Fw: TGAGATCCGGCTGCTAACAAAGCCCGAAAGG and Rv: CTCTGTCAAGATAGCATCCTTAGGTGCTTCAACGAACG. The construct was then transformed into E. coli BL21(DE3) competent cell and the clones were confirmed by DNA sequencing.
Protein expression and purification
Proteins were produced in batch cultivations in E. coli BL21(DE3) in LB-medium with 25 mg/L kanamycin as antibiotic. Gene expression in the cells was induced with IPTG in the mid-log phase. The cells were recovered by centrifugation at 5000 × g for 10 min and washed once in binding buffer (sodium phosphate buffer pH 7.4) and then re-suspended in the same buffer. Cells were lysed by ultrasonication on ice 3 × 3 min in 3 min intervals and then centrifuged at 26700 × g for 20 min. Supernatants (containing His-tagged proteins) were purified by immobilized metal ion affinity chromatography 0MAC) (AKTA Prime, Uppsala, Sweden). Imidazole was used to elute bound protein. After elution, the proteins were dialyzed (3.5 kDa cutoff, SpectrumLab, CA) in 20 mM sodium phosphate buffer, pH 7.4, and 0.5 M NaCl at 4°C overnight and the following day by changing the dialysis buffer once. Purity of the protein solutions was determined by SDS–PAGE (Bio-Rad, Sweden) with 12.5% acrylamide. Protein concentration after purification was estimated spectrophotometrically at 280 nm. Theoretically calculated extinction coefficients were obtained using ProtParam (www.expasy.org) and were 149,660 M/cm (full-length enzyme) and 82,850 M/cm (catalytic domain).
Molecular mass determination
The molecular masses of IMAC purified β-xylosidase complexes were determined by SEC according to Michlmayr et al. (2013) using a column of 110 mL. All other conditions were as described by Michlmayr et al. (2013) including the calibration standard that was bought from Sigma-Aldrich. The linear flow was set at 0.1 mL/cm.
Standard β-xylosidase assay
A standard assay was performed to monitor activity of the protein during the production/purification process. The assay was performed in 0.5 mL reactions with 1 mM p-nitrophenyl-β-d-xylopyranoside in 50 mM sodium phosphate buffer, pH 7.0, at 37°C for 1 min and the reaction was stopped using 1 volume of 0.5 M sodium carbonate (Merck). The absorbance was measured at 400 nm in a BioWave2 spectrophotometer. The extinction coefficient (18300 M/cm) of released p-nitrophenol (pNP) was determined using a standard of pNP (Merck).
Specific activity for p-nitrophenyl-glycosides
Specific activity of p-nitrophenyl-glycosides was done in a 96-well plate (Corning, Vordingborg, Denmark) in 200 µL with 1 mM substrate concentration in 50 mM citrate phosphate buffer (CPB) at pH 6.0 at 37°C. Absorbance was measured continuously for 1 min at 400 nm in a microplate reader (Multiscan GO, Thermo Scientific). The extinction coefficient (2970 M/cm) of released pNP at pH 6.0 at 37°C was determined using the same standard as above.
Specific activity for oligosaccharide substrates
Reactions were performed in 200 µL with oligosaccharides (X2–X4) at a concentration of 1 mM using 50 mM CPB at pH 6.0 and 37°C. Samples were withdrawn after 1, 2 and 3 min and the reaction stopped by incubating the samples at 95°C for 5 min. Release of monosaccharides was measured by HPAEC-PAD using the conditions described below (see Kinetic parameters).
pH and temperature surface plot
Based on the initial substrate screening, a surface plot of the optimum pH and temperature was done in 125 µL reactions using 1 mM p-nitrophenyl-β-d-xylopyranoside and the reaction was stopped using in 1 volume of 0.5 M sodium carbonate after 30, 60 and 90 s. CPB buffers were prepared in the range of pH 4.0–7.5 and the reactions were performed at 30, 37, 45, 55 and 65°C. Absorbance of 200 µL reaction mixes was measured in a 96-well plate at 400 nm in the microplate reader described above using the extinction coefficient of 18,300 M/cm and a path length of 0.58 cm.
Temperature stability
Remaining activity was determined after 10, 20, 40 and 60 min by incubating the protein at 30, 37, 45 and 55°C. The specific activity was assayed as before except that the concentration of p-nitrophenyl-β-d-xylopyranoside was 5 mM.
Kinetic parameters
All kinetic experiments were determined at the same assay conditions, 37°C and 50 mM CPB at pH 6.0. Initial rates were measured in the range of 0.5–60 mM of p-nitrophenyl-β-d-xylopyranoside using 2 µg/mL of protein in 200 µL reactions. Absorbance was measured continuously for 1 min at 400 nm in a microplate reader at 37°C (2970 M/cm). Natural substrate kinetics was performed in the range of 0.5–25 mM using 0.1 µg/mL of protein in 50 µL reactions for 5 min and the reaction was stopped by incubating the samples at 95°C for 5 min. Release of xylose was measured by HPAEC-PAD (Dionex, CA) with a 250 × 3 mm i.d. 5.5 µm CarboPac PA200 and a guard column 50 × 3 mm of the same material with a mobile phase (0.5 mL/min) of constant 100 mM NaOH (Merck) and a gradient of sodium acetate (Sigma). Values of KM and kcat were calculated by nonlinear regression to the Michaelis–Menten equation using GraphPad Prism6 built in functions.
Homology modelling
The three-dimensional structural models of the full-length protein were obtained by homology modeling, using the YASARA software (http://www.yasara.org). The accuracy of the models obtained is supported by: (i) the relatively high identity in amino acid sequences, (ii) the molecular mechanics calculations including explicit solvent with molecules of water and (iii) the use of several crystallographic templates for generating the hybrid models, which in this case have higher Z-scores than using only one template.
The target sequence contained 583 amino acids, of which the first 16 residues were excluded (including the 6× His tag used for purification). Two models were predicted, the first one as a single protomer and the second one as a homo-tetramer. The chosen modeling parameters are listed in Supplementary data, Table SI. The program built the model according to the alignment of the target sequence with several PDB templates from crystallographic structures. The templates were the crystallographic structures of β-xylosidases from three bacterial species: S ruminantium, G. stearothermophilus and B. halodurans (Table III). The 3D hybrid models obtained showed the lowest root mean square deviation (RMSD) to the G. stearothermophilus crystallographic structure (PDB 2EXI), despite lower overall sequence similarity (Table III). The overall RMSD of the atomic coordinates of the alpha carbons (Cα) was 0.896 Å when the hybrid model was superimposed with 2EXI. Details of the contribution of each template to the hybrid model are summarized in Supplementary data, Tables SII and SIII. The modeled loops were optimized and the side-chains fine-tuned by combining steepest descent and simulated annealing strategies for minimization. Half of the refined models were evaluated by quality Z-scores, which indicate how many standard deviations the model quality is away from the average high-resolution X-ray structure. Subsequently, a full unrestrained simulated annealing minimization was run for each model. Finally, to increase the accuracy, the best parts of the models generated were combined to obtain a hybrid model.
The atomic coordinates of xylobiose (X2) were transferred from the crystallographic complex xylobiose-XynB3 (PDB: 2EXJ) from G. stearothermophilus (Brüx et al. 2006) to the corresponding binding site in the model of WXyn43. After simulated annealing calculations of the resulting complex in explicit solvent with molecules of water, the interactions between the ligand and the protein model were compared with those of the crystal structure.
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Conflict of interest statement
None declared.
Funding
This work was supported by VINNOVA, Lund University: Antidiabetic Food Centre (VINN Excellence Centre) and by the Swedish Research Council (VR). Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council.
Abbreviations
AOS, arabinooligosaccharides; AXOS, arabinoxylooligosaccharides; CPB, citrate phosphate buffer; DP, degree of polymerization; GHs, glycoside hydrolases; LAB, lactic acid bacteria; LB, Luria Bertani; pNP, p-nitrophenol; pNPAf, p-nitrophenyl-α-l-arabinofuranoside; pNPX, p-nitrophenyl-β-d-xylopyranoside; SEC, size exclusion chromatography; XOS, xylooligosaccharides.
Supplementary Material
References
- Aachary AA, Prapulla SG. 2011. Xylooligosaccharides (XOS) as an emerging prebiotic: Microbial synthesis, utilization, structural characterization, bioactive properties, and applications. Compr Rev Food Sci Food Saf. 10:2–16. [Google Scholar]
- Amaretti A, Bernardi T, Leonardi A, Raimondi S, Zanoni S, Rossi M. 2013. Fermentation of xylo-oligosaccharides by Bifidobacterium adolescentis DSMZ 18350: Kinetics, metabolism, and beta-xylosidase activities. Appl Microbiol Biotechnol. 97:3109–3117. [DOI] [PubMed] [Google Scholar]
- Amari M, Laguerre S, Vuillemin M, Robert H, Loux V, Klopp C, Morel S, Gabriel B, Remaud-Siméon M, Gabriel V et al. . 2012. Genome sequence of Weissella confusa LBAE C39–2, isolated from a wheat sourdough. J Bacteriol. 194:1608–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert WF, Courtin CM, Verbeke K, Van de Wiele T, Verstraete W, Delcour JA. 2011. Prebiotic and other health-related effects of cereal-derived arabinoxylans, arabinoxylan-oligosaccharides, and xylooligosaccharides. Crit Rev Food Sci Nutr. 51:178–194. [DOI] [PubMed] [Google Scholar]
- Brunzelle JS, Jordan DB, McCaslin DR, Olczak A, Wawrzak Z. 2008. Structure of the two-subsite b-D-xylosidase from Selenomonas ruminantium in complex with 1,3-bis[tris(hydroxymethyl)methylamino]propane. Arch Biochem Biophys. 474:157–166. [DOI] [PubMed] [Google Scholar]
- Brüx C, Ben-David A, Shallom-Shezifi D, Leon M, Niefind K, Shoham G, Shoham Y, Schomburg D. 2006. The structure of an inverting GH43 beta-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J Mol Biol. 359:97–109. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: An integrative view. Cell. 148:1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden R, Karppinen S, Ojanen S, Tenkanen M, Fagerstrom R, Matto J, Saarela M, Mattila-Sandholm T, Poutanen K. 2002. In vitro fermentation of cereal dietary fibre carbohydrates by probiotic and intestinal bacteria. J Sci Food Agric. 82:781–789. [Google Scholar]
- Falck P, Precha-Atsawanan S, Grey C, Immerzeel P, Stålbrand H, Adlercreutz P, Nordberg Karlsson E. 2013. Xylooligosaccharides from hardwood and cereal xylans produced by a thermostable xylanase as carbon sources for Lactobacillus brevis and Bifidobacterium adolescentis. J Agric Food Chem. 61:7333–7340. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 3:289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullon P, Moura P, Esteves MP, Gtrio FM, Dominguez H, Parajo JC. 2008. Assessment on the fermentability of xylooligosaccharides from rice husks by probiotic bacteria. J Agric Food Chem. 56:7482–7487. [DOI] [PubMed] [Google Scholar]
- Hijova E, Chmelarova A. 2007. Short chain fatty acids and colonic health. Bratislava Med J Bratisl Lek Listy. 108:354–358. [PubMed] [Google Scholar]
- Immerzeel P, Falck P, Galbe M, Adlercreutz P, Nordberg Karlsson E, Stålbrand H. 2014. Extraction of water-soluble xylan from wheat bran and utilization of enzymatically produced xylooligosaccharides by Lactobacillus, Bifidobacterium and Weissella spp. LWT-Food Sci Technol. 56:321–327. [Google Scholar]
- Jordan D, Wagschal K, Grigorescu A, Braker J. 2013. Highly active β-xylosidases of glycoside hydrolase family 43 operating on natural and artificial substrates. Appl Microbiol Biotechnol. 97:4415–4428. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Vaughan EE. 2009. Probiotic and gut lactobacilli and bifidobacteria: Molecular approaches to study diversity and activity. Annu Rev Microbiol. 63:269–290. [DOI] [PubMed] [Google Scholar]
- Lagaert S, Pollet A, Delcour JA, Lavigne R, Courtin CM, Volckaert G. 2011. Characterization of two beta-xylosidases from Bifidobacterium adolescentis and their contribution to the hydrolysis of prebiotic xylooligosaccharides. Appl Microbiol Biotechnol. 92:1179–1185. [DOI] [PubMed] [Google Scholar]
- Lagaert S, Pollet A, Delcour JA, Lavigne R, Courtin CM, Volckaert G. 2010. Substrate specificity of three recombinant alpha-L-arabinofuranosidases from Bifidobacterium adolescentis and their divergent action on arabinoxylan and arabinoxylan oligosaccharides. Biochem Biophys Res Commun. 402:644–650. [DOI] [PubMed] [Google Scholar]
- Lagaert S, Van Campenhout S, Pollet A, Bourgois TM, Delcour JA, Courtin CM, Volckaert G. 2007. Recombinant expression and characterization of a reducing-end xylose-releasing exo-oligoxylanase from Bifidobacterium adolescentis. Appl Environ Microbiol. 73:5374–5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Park JY, Jeong HR, Heo HJ, Han NS, Kim JH. 2012. Probiotic properties of Weissella strains isolated from human faeces. Anaerobe. 18:96–102. [DOI] [PubMed] [Google Scholar]
- Michlmayr H, Hell J, Lorenz C, Böhmdorfer S, Rosenau T, Kneifel W. 2013. Arabinoxylan oligosaccharide hydrolysis by family 43 and 51 glycosidases from Lactobacillus brevis DSM 20054. Appl Environ Microbiol. 79:6747–6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moure A, Gullon P, Dominguez H, Parajo JC. 2006. Advances in the manufacture, purification and applications of xylo-oligosaccharides as food additives and nutraceuticals. Process Biochem. 41:1913–1923. [Google Scholar]
- Nurizzo D, Turkenburg JP, Charnock SJ, Roberts SM, Dodson EJ, McKie VA, Taylor EJ, Gilbert HJ, Davies GJ. 2002. Cellvibrio japonicus α-L-arabinanase 43A has a novel five-blade β-propeller fold. Nat Struct Mol Biol. 9:665–668. [DOI] [PubMed] [Google Scholar]
- Ohland CL, MacNaughton WK. 2010. Probiotic bacteria and intestinal epithelial barrier function. Am J Physiol Gastrointest Liver Physiol. 298:G807–G819. [DOI] [PubMed] [Google Scholar]
- Patel A, Falck P, Shah N, Immerzeel P, Adlercreutz P, Stålbrand H, Prajapati JB, Holst O, Nordberg Karlsson E. 2013. Evidence for xylooligosaccharide utilization in Weissella strains isolated from Indian fermented foods and vegetables. FEMS Microbiol Lett. 346:20–28. [DOI] [PubMed] [Google Scholar]
- Patel A, Lindström C, Patel A, Prajapati J, Holst O. 2012. Probiotic properties of exopolysaccharide producing lactic acid bacteria isolated from vegetables and traditional Indian fermented foods. Int J Fermented Foods. 1:87–101. [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera-A visualization system for exploratory research and analysis. J Comput Chem. 13:1605–1612. [DOI] [PubMed] [Google Scholar]
- Rastall R. 2010. Functional oligosaccharides: Application and manufacture. Annu Rev Food Sci Technol. 1:305–339. [DOI] [PubMed] [Google Scholar]
- Roberfroid M. 2007. Prebiotics: The concept revisited. J Nutr. 137:830S–837S. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LCM, Finlay BB. 2010. Gut microbiota in health and disease. Physiol Rev. 90:859–904. [DOI] [PubMed] [Google Scholar]
- van Den Broek LAM, Voragen AGJ. 2008. Bifidobacterium glycoside hydrolases and (potential) prebiotics. Innov Food Sci Emerging Technol. 9:401–407. [Google Scholar]
- Wallace TC, Guarner F, Madsen K, Cabana MD, Gibson G, Hentges E, Sanders ME. 2011. Human gut microbiota and its relationship to health and disease. Nutr Rev. 69:392–403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.