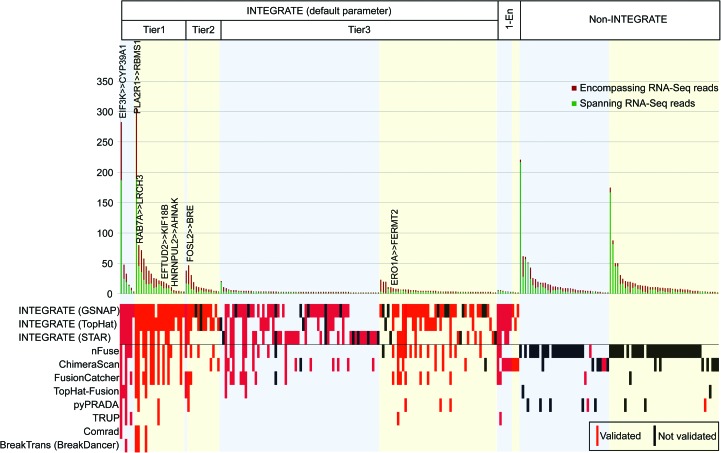
Figure 2.
Gene fusion validation. Targeted cDNA-capture validation was attempted for 240 gene fusion candidates called by INTEGRATE and eight additional gene fusion detection methods, resulting in the validation of 138 gene fusions. Gene fusion candidates nominated by INTEGRATE using default parameters and a threshold of one encompassing RNA-seq read (1-En) are shown on the left, whereas candidates nominated by eight additional programs, and not INTEGRATE (“non-INTEGRATE”), are shown on the right. See Supplemental Figure 2 for tiers of INTEGRATE. In each category, gene fusion candidates are further divided into rearrangement classes: inter-chromosomal (blue shade) and intra-chromosomal (yellow shade), and sorted in descending order of total RNA-seq read support (dark red bar—encompassing RNA-seq reads; green bar—spanning RNA-seq reads). Previously reported gene fusions are written at the top of the bars. In the lower panel, each row corresponds to the gene fusion candidates nominated by each program. INTEGRATE, Comrad, BreakTrans, and nFuse use both RNA-seq and WGS data. INTEGRATE is shown using the RNA-seq alignments from GSNAP, TopHat2, and STAR, separately. ChimeraScan, Tophat-Fusion, FusionCatcher, pyPRADA, and TRUP use only RNA-seq data. Red boxes indicate nominated gene fusions that were experimentally validated, black boxes indicate nominated gene fusions that did not have validation read support, and the lack of a red or black box indicates an algorithm did not nominate the gene fusion candidate.