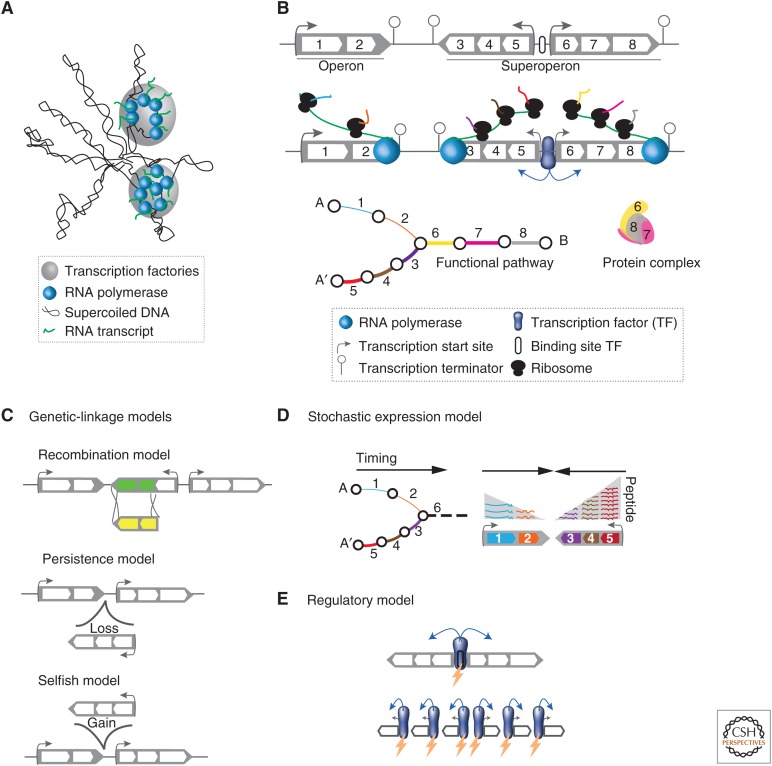
Figure 3.
The genetic organization of gene expression. (A) Schematic representation of the transcription factory model, in which multiple active RNA polymerases are concentrated at discrete sites in the nucleoid. (B) Bacterial genes are organized into operons, which group in superoperons. In addition to being physically close in the genome, genes in operons are cotranscribed, coregulated, and encode proteins involved in the same functional pathway or protein complex. Superoperons are not cotranscribed, but may share regulatory regions. (C) Schematic representation of three models aiming at explaining the formation and conservation of operons based on genetic linkage. (D) Transcription of genes in a single transcript is expected to diminish gene expression noise and ensure more precise stoichiometry. It also allows responding optimally to demand for a given pathway when the first genes to be transcribed are those starting the functional pathway. (E) Operons place several genes under the same regulatory region that is thus subject to more efficient selection.