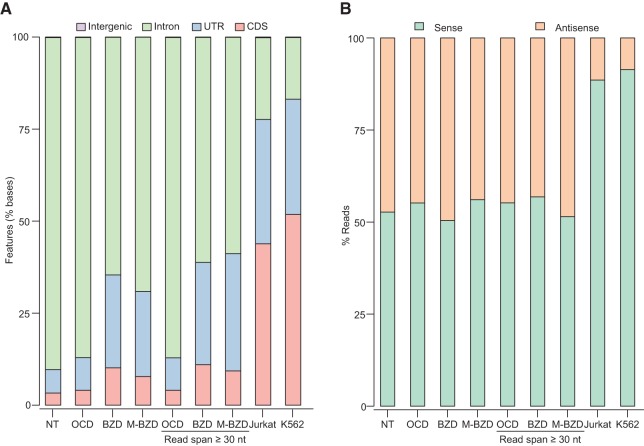
FIGURE 4.
Human plasma RNA is enriched in intron and antisense sequences compared with whole-cell RNAs. Reads mapping to protein-coding genes were analyzed to assess coverage across different regions and both DNA strands in RNA-seq data sets constructed with TGIRT enzymes for total plasma or whole-cell RNA prepared and treated in different ways. These include plasma RNA prepared by the Direct-zol method with no further treatment (NT; combined DS1–3), after on-column DNase I treatment (OCD; combined DS7–10), or after Baseline-ZERO DNase treatment (BZD; DS11); plasma RNA prepared by the mirVana combined method after Baseline-ZERO DNase treatment (M-BZD; DS16); and ribo-depleted and fragmented whole-cell RNA from Jurkat cells (TeI4c RT; DS18) or K562 cells (GsI-IIC RT; DS19). (A) Stacked bar graphs showing the percentage of bases in protein-coding gene reads that mapped to coding sequences (CDS), introns, 5′- and 3′-untranslated regions (UTRs), and intergenic regions. (B) Stacked bar graphs showing the proportion of concordant read pairs that mapped to the sense and antisense strands of protein-coding genes. In A and B, reads that mapped to protein-coding genes were filtered to remove those with >50% of the read length overlapping embedded small ncRNAs, and the percentage of bases or reads mapping to different regions or strands was calculated by using picard tools. Reads from the OCD, BZD, and M-BZD data sets were analyzed with or without removal of read pairs with a span of <30 nt to exclude short DNA fragments that may have escaped DNase treatment.