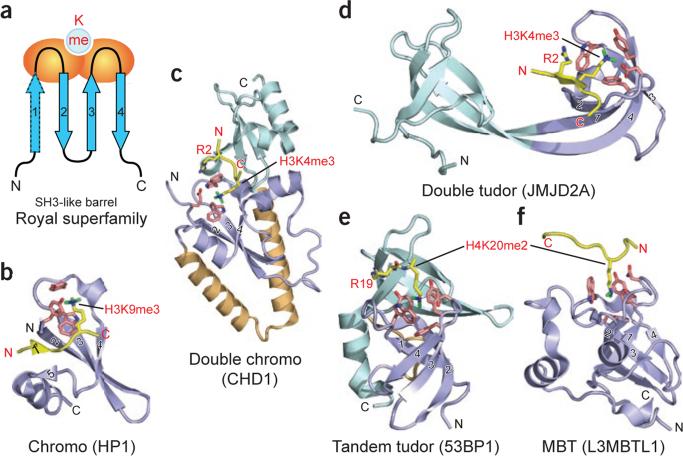
Figure 3.
Readout of methyllysine marks by Royal-superfamily modules. (a) Topology of the Royal-superfamily fold. Blue arrows, β-strands that form an incomplete β-barrel reminiscent of the SH3 domain fold; orange ovals, loops participating in methyllysine reader pocket formation; light blue circle, binding pocket at one end of the module. Classical chromo modules have only three core β-strands (labeled 2–4) and one orphaned extra strand (labeled 5). Upon complex formation, the histone peptide completes this five-stranded β-barrel fold by introducing an extra β-strand at position 1 prime, sandwiched between strands 2 and 5 (see b). This sandwiching binding mode occurs mainly with chromodomains. In other Royal superfamily members, the interactions are more varied; however, docking in the β-strand conformation to extend one edge of an existing β-sheet is not uncommon (see d). (b–f) Examples of known complex structures in the Royal superfamily, ranging from higher methylation state–specific readers (b–d) to lower methylation state–specific readers (e and f). Strands that form the SH3-like β-barrel are in slate, numbered as in a. Coordinates have PDB codes 1KNE (b), 2B2W (c), 2GFA (d), 2IG0 (e) and 2PQW (f).