Background: Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is the essential enzyme for carbon fixation.
Results: Rhodospirillum rubrum requires Rubisco to metabolize 5-methylthioadenosine and carbon dioxide as sulfur and carbon source, respectively.
Conclusion: Rubisco concurrently catalyzes reactions for distinct carbon and sulfur metabolic pathways under anaerobic conditions.
Significance: A novel role of Rubisco in sulfur metabolism provides insight into Rubisco catalytic versatility.
Keywords: carbon fixation; enzyme; metabolism; metabolomics; ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO); sulfur; metabololomics; methylthioadenosine; sulfur salvage; 5-methylthioadenosine
Abstract
All organisms possess fundamental metabolic pathways to ensure that needed carbon and sulfur compounds are provided to the cell in the proper chemical form and oxidation state. For most organisms capable of using CO2 as sole source of carbon, ribulose-1,5-bisphosphate (RuBP) carboxylase/oxygenase (Rubisco) catalyzes primary carbon dioxide assimilation. In addition, sulfur salvage pathways are necessary to ensure that key sulfur-containing compounds are both available and, where necessary, detoxified in the cell. Using knock-out mutations and metabolomics in the bacterium Rhodospirillum rubrum, we show here that Rubisco concurrently catalyzes key and essential reactions for seemingly unrelated but physiologically essential central carbon and sulfur salvage metabolic pathways of the cell. In this study, complementation and mutagenesis studies indicated that representatives of all known extant functional Rubisco forms found in nature are capable of simultaneously catalyzing reactions required for both CO2-dependent growth as well as growth using 5-methylthioadenosine as sole sulfur source under anaerobic photosynthetic conditions. Moreover, specific inactivation of the CO2 fixation reaction did not affect the ability of Rubisco to support anaerobic 5-methylthioadenosine metabolism, suggesting that the active site of Rubisco has evolved to ensure that this enzyme maintains both key functions. Thus, despite the coevolution of both functions, the active site of this protein may be differentially modified to affect only one of its key functions.
Introduction
Rubisco is the key enzyme of the Calvin-Benson-Bassham (CBB)3 reductive pentose phosphate pathway and is thought to be the most abundant protein on earth, responsible for the bulk of biologically produced organic carbon. Over 60 years of discovery and experimentation with Rubisco have provided much insight as to the mechanism of catalysis (1), the enzyme's physiological relevance in both CO2 and O2 metabolism (2), and aspects of protein folding and assembly dynamics (3).
Based on amino acid sequence homologies, three forms of Rubisco have been described (forms I, II, and III) (4) that are capable of catalyzing the typical carboxylase reaction required for CO2 fixation via the CBB pathway. Some 17 years ago, a new member of the Rubisco family was discovered, the Rubisco-like protein (RLP), or form IV Rubisco (5). RLPs lack the capacity to catalyze the typical carboxylation reaction and have been identified in proteobacteria, cyanobacteria, archaea, and algae (6, 7). No functional similarity was initially found between Rubisco and RLP due to the substitution of several key active site residues in the latter (7); indeed in Rhodospirillum rubrum, 7 of the 19 required active site residues of its form II Rubisco are altered in the R. rubrum RLP. However, significant structural homology exists between the two proteins (8). Moreover, the RLP from Chlorobaculum (Chlorobium) tepidum was found to be involved in sulfur metabolism (5, 9), and subsequent studies with RLP from Bacillus subtilis (10) and other organisms (11, 12) indicated that the YkrW class of RLPs catalyzes a key reaction of an essential sulfur (methionine) salvage pathway, which enables these organisms to metabolize and detoxify MTA (see Fig. 1; compound 4). They do this by catalyzing the conversion of 2,3-diketo-5-methylthiopentyl-1-phosphate (DK-MTP-1-P; 10) to 2-hydroxy-3-keto-5-methylthiopent(1)ene-1-phosphate (HK-MTPene-1P; 11) (see Fig. 1; reaction L) (10, 12). Mechanistically, this reaction is quite analogous to the enolization of ribulose-1,5-bisphosphate (RuBP) catalyzed by Rubisco. Not all classes of RLP catalyze this reaction, however, as the class IV photo RLP from R. rubrum was found to catalyze a unique isomerization reaction using 5-methylthioribulose-1-phosphate (MTRu-1P; 7) as substrate to produce 1-methylthio-xylulose-5-phosphate (MTXu-5P; 8a) (see Fig. 1; H). As such, this RLP appears to be part of a novel and hitherto undescribed sulfur salvage pathway linked to isoprenoid biosynthesis under aerobic growth conditions (13).
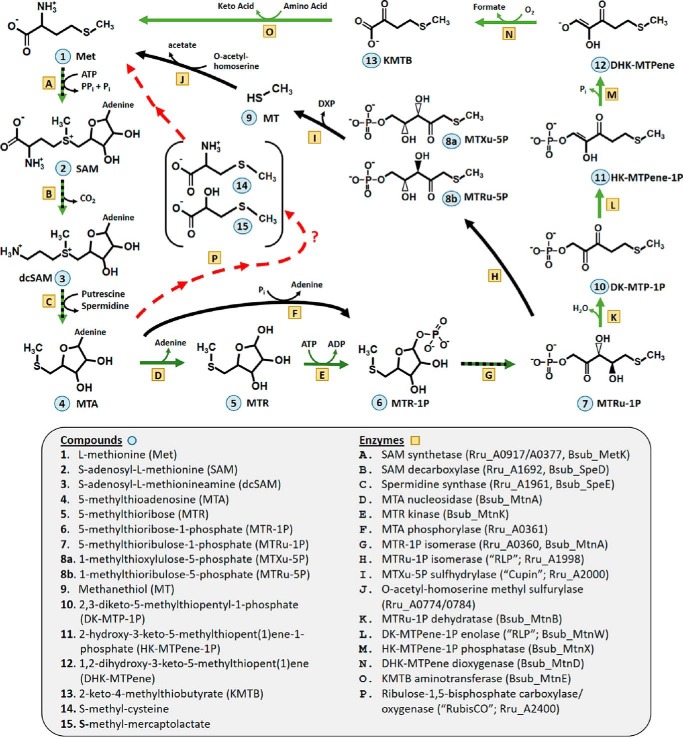
FIGURE 1.
MTA metabolism pathways for sulfur (methionine) salvage (10, 13) in R. rubrum (aerobic pathway in black, anaerobic in red) and B. subtilis (aerobic pathway in green, shared in black/green). Under anaerobic conditions, initially R. rubrum metabolizes MTA (4) to MTR-1P (6) using the bifunctional MTA phosphorylase (F), whereas B. subtilis employs two separate enzymes (D and E) with MTR (5) as an intermediate. Subsequently, after formation of MTRu-1P (7) by MTR-1P isomerase (G), the pathways diverge, wherein B. subtilis and R. rubrum RLP catalyze separate and distinct enolase (L) (10) and isomerase (H) (27) reactions, respectively. The R. rubrum RLP product, MTXu-5P (8a), is metabolized via a cupin-like MTXu-5P sulfhydrylase (I) into MT (9) and DXP, the latter of which proceeds to isoprenoid biosynthesis (13). In R. rubrum, under anaerobic conditions, RubisCO (P) catalyzes an unknown but required reaction for MTA metabolism that subsequently leads to the production of S-methyl-cysteine (14) and S-methyl-mercaptolactate (15). Compounds and enzymes involved in the pathway are designated by numerals and letters, respectively.
Despite the obvious functional differences between Rubisco and RLP, the conservation of structural features and the identity of several key active site residues and motifs (6–8) suggested that Rubisco and RLP might still have some unknown functional similarity. R. rubrum provides an interesting system to study the roles of RLP and Rubisco as this organism synthesizes both of these proteins. In support of the notion that Rubisco and RLP might share some functional similarity, previous studies (14) indicated that R. rubrum Rubisco and RLP were both required for MTA-dependent growth of R. rubrum. However, Rubisco was required under anaerobic photosynthetic conditions, whereas RLP was required under aerobic (non-photosynthetic) growth conditions. Contrary to an earlier study (10), R. rubrum Rubisco does not catalyze the typical DK-MTP-1P to HK-MTPene-1P enolization reaction of the B. subtilis type sulfur salvage pathway (15) (see Fig. 1; L).
In the current study, we show through in vivo experiments that representatives of all bona fide Rubisco forms (I, II, and III) catalyze key reactions for both CO2 fixation and MTA metabolism. Complete inactivation of the CO2 fixation reaction does not prevent or affect Rubisco-dependent MTA metabolism. In addition, knock-out metabolomics revealed that Rubisco's role in anaerobic MTA metabolism is potentially linked to S-methyl-cysteine and S-methyl-mercaptolactate production. These results suggest that the quintessential carbon fixation enzyme, Rubisco, also catalyzes an essential reaction required for anaerobic MTA metabolism.
Experimental Procedures
Bacterial Strains
Wild type R. rubrum Str-2 is a spontaneous streptomycin-resistant derivative of strain S1 (ATCC 11170) (16). Strain I19A (ΔcbbM) is a derivative of the WT strain in which the Rubisco gene (cbbM) is disrupted by a kanamycin marker (17). Strain IR (ΔcbbM/ΔrlpA) is a derivative of I19A in which the RLP gene (rlpA) is disrupted by a gentamycin marker (14). Strain I19NifA (ΔcbbM,nifA-M173V) is a derivative of I19A containing a spontaneous point mutation (M173V) in the nifA gene (18). Inactivation of the RLP gene (rlpA) in strain I19NifA by insertion of a gentamycin marker to create strain IRNifA (ΔcbbM/ΔrlpA,nifA-M173V) was performed exactly as described previously (14). Rhodobacter capsulatus strain SB I/II− is a derivative of wild type strain SB1003 in which both Rubisco form I (cbbLS) and form II (cbbM) genes are disrupted by spectinomycin and kanamycin markers, respectively, as described previously (19).
Plasmids for Complementation Studies
plasmid pRPS-MCS3 (19) is composed of the broad spectrum host plasmid pBBR1-MCS3 containing the R. rubrum cbb promoter and cbbR regulatory gene for expression of Rubisco genes in trans in the R. rubrum host strains. Plasmids pRPS-6301, pRPS-6301-F97L, and pRPS-6301-D103V were constructed by cloning Synechococcus sp. 6301 form I Rubisco large and small subunit genes (rbcLS) into pRPS-MCS3 without or with the large subunit mutations F97L and D103V, respectively, as described previously (20). The form III Rubisco gene (rcbL) of Methanococcoides burtonii strain DSM6242 was cloned from genomic DNA (gift of Dr. Kevin Sowers) into pCR-BluntII-TOPO (Invitrogen) using primers (ATGAGTTTAATCTATGAGG and TTATCTATTCAAATAGAACTC), followed by subcloning into PstI- and XbaI- (New England Biolabs) digested pRPS-MCS3 using the In-Fusion cloning system (Clontech) and primers (TTGATATCGAATTCCTGCAGATGAGTTTAATCTATGAGGACCTGGTAAAATCG and TGGCGGCCGCTCTAGATTATCTATTCAAATAGAACTCGATCGCTTCTGC) to create plasmid pRPS-MBR (21). Plasmids pRPS-Rrub-cbbM and pRPS-Rpal-cbbM contain the wild type R. rubrum and Rhodopseudomonas palustris form II Rubisco gene (cbbM), respectively, cloned into pRPS-MCS3 as described previously (14, 22). Plasmids pJG336 and pJG106 are composed of a 20-kb Rhodobacter sphaeroides HindIII fragment containing the form I Rubisco genes (cbbLS) and form II Rubisco gene (cbbM), respectively, cloned into plasmid pVK102 (23).
Bacterial Growth and Complementation Studies
Bacterial conjugation for complementation studies was performed by bi-parental mating as described previously (14, 17), using Escherichia coli strain SM-10 as the donor strain. All plasmids employed contained a tetracycline resistance marker that was used for counter-selection of transconjugants. All R. rubrum IR and IRNifA transconjugants were initially grown under anaerobic, photoheterotrophic conditions in Ormerod's minimal medium (24) supplemented with 20 mm dl-malate (Sigma), 50 μg/ml streptomycin, 25 μg/ml kanamycin, 15 μg/ml gentamycin, and 10 μg/ml tetracycline. All R. capsulatus SBI/II− transconjugants were initially grown under aerobic, chemoheterotrophic conditions in PYE (3 g/liter peptone, 3 g/liter yeast extract, 1× Ormerod's basal salts (24), 1 μg/ml nicotinic acid, 1 μg/ml thiamine, 15 μg/ml biotin) supplemented with 25 μg/ml spectinomycin, 25 μg/ml kanamycin, and 2 μg/ml tetracycline.
Cells were harvested at mid to late log phase by centrifugation and washed three times in Ormerod's minimal medium depleted of sulfur by substituting all sulfate-containing salts with their chloride or acetate analog. Cells were then inoculated into anaerobic culture tubes containing 10 ml of sulfur-free minimal medium supplemented ammonium sulfate (Amresco), MTA (Sigma), and l-methionine (Sigma), where indicated. Tubes were capped, sealed, and grown anaerobically at 30 °C either photoheterotrophically with 20 mm dl-malate as sole carbon source under a 5% H2/95% N2 atmosphere or photoautotrophically with CO2 as sole carbon source under a 5% CO2/95% H2 atmosphere, where indicated. Bacterial growth was monitored at OD660 nm, and bacterial growth curves were fit by non-linear weighted regression (MATLAB, MathWorks) to a sigmoidal-logistic model (25) to measure the growth rate (Table 1).
TABLE 1.
Growth phenotypes of strains
Shown are R. rubrum (R. rub) and R. capsulatus (R. caps) strains and corresponding plasmids used in this study with strain/plasmid references in parentheses. Anaerobic phototrophic growth phenotypes are shown with given carbon/sulfur sources indicated by (+) growth, (−/+) very slow growth, (−) no growth. For (+) and (−/+) phenotypes, doubling time, T, is given in hours. NA, not applicable; R. sph, R. sphaeroides; M. bur, M. burtonii; R. pal, R. palustris; Synec, Synechococcus.
| Strain | Plasmid | Carbon | Sulfur | Growth | T |
|---|---|---|---|---|---|
| h | |||||
| R. rub WT (16) | None | Malate | SO4 | + | 11 ± 2 |
| Malate | MTA | + | 16 ± 3 | ||
| CO2 | SO4 | + | 38 ± 4 | ||
| CO2 | Met | + | 45 ± 5 | ||
| CO2 | MTA | + | 23 ± 3 | ||
| R. rub WR (14) | None | Malate | SO4 | + | 15 ± 2 |
| Malate | MTA | + | 18 ± 3 | ||
| CO2 | SO4 | + | 46 ± 6 | ||
| CO2 | Met | + | 51 ± 8 | ||
| CO2 | MTA | + | 20 ± 2 | ||
| R. rub I19A (17) | None | Malate | SO4 | + | 18 ± 4 |
| Malate | MTA | − | NA | ||
| CO2 | SO4 | − | NA | ||
| CO2 | Met | − | NA | ||
| CO2 | MTA | − | NA | ||
| R. rub I19NifA (18) | None | Malate | SO4 | + | 19 ± 2 |
| Malate | MTA | − | NA | ||
| R. rub IRNifA (this study) | None | Malate | SO4 | + | 19 ± 2 |
| Malate | MTA | −/+ | 100 ± 30 | ||
| pRPS-MCS3 (19) | Malate | SO4 | + | 19 ± 4 | |
| Malate | MTA | −/+ | 97 ± 24 | ||
| pRPS-R. rub-cbbM (14) | Malate | SO4 | + | 21 ± 2 | |
| Malate | MTA | + | 38 ± 2 | ||
| pRPS-R. pal-cbbM (22) | Malate | SO4 | + | 36 ± 3 | |
| Malate | MTA | + | 22 ± 2 | ||
| R. rub IR (14) | None | Malate | SO4 | + | 21 ± 2 |
| Malate | MTA | −/+ | 130 ± 45 | ||
| CO2 | SO4 | − | NA | ||
| CO2 | Met | − | NA | ||
| CO2 | MTA | − | NA | ||
| pJG336 (R. sph cbbLS) (23) | Malate | SO4 | + | 20 ± 5 | |
| Malate | MTA | + | 19 ± 10 | ||
| pJG106 (R. sph cbbM) (23) | Malate | SO4 | + | 55 ± 7 | |
| Malate | MTA | + | 59 ± 14 | ||
| pRPS-MBR (M. bur rbcL) (this study) | Malate | SO4 | + | 15 ± 2 | |
| Malate | MTA | + | 18 ± 2 | ||
| R. rub IR (14) | pRPS-6301 (Synec rbcLS) (20) | Malate | MTA | + | 52 ± 14 |
| pRPS-6301-F97L | Malate | MTA | + | 25 ± 6 | |
| pRPS-6301-D103V | Malate | MTA | + | 52 ± 21 | |
| R. caps SBI/II- (19) | pRPS-6301 (Synec rbcLS) (20) | CO2 | SO4 | + | 26 ± 7 |
| pRPS-6301-F97L | CO2 | SO4 | − | NA | |
| pRPS-6301-D103V | CO2 | SO4 | − | NA |
Immunoblot Assay
R. rubrum cells from growth studies were harvested at early stationary phase by centrifugation, resuspended in 500 μl of TEM buffer (50 mm Tris-Cl, pH 7.5, 1 mm EDTA, and 1 mm β-mercaptoethanol), disrupted by sonication, and centrifuged. Soluble proteins were resolved by 12% SDS-PAGE and transferred to Immobilon-P membrane (Millipore) according to the manufacturer's instructions. Primary antiserum was raised against R. rubrum Rubisco holoenzyme and was used at a dilution of 1:3,000. Alkaline phosphatase-labeled goat anti-rabbit IgG (Bio-Rad Laboratories) was used as the secondary antibody and detected by the enhanced chemiluminescence method (GE Healthcare).
Metabolite Detection
R. rubrum strains were grown to late log phase under anaerobic, photoheterotrophic conditions in Ormerod's malate minimal medium supplemented with 1 mm MTA (strains WT and WR) or 1 mm sulfate (strain IR), harvested by centrifugation, and washed three times with sulfur-free medium under strict anaerobic conditions. Cells were resuspended in Ormerod's malate minimal medium supplemented with 1 mm MTA or no sulfur source and incubated under anaerobic, photoheterotrophic conditions for the designated amount of time (see Fig. 6) and harvested by centrifugation. Metabolites were extracted and identified by LC-Fourier transform MS as described previously (13) with the exception that the detector was an LTQ-Velos Pro Orbitrap (Thermo-Fisher Scientific) instead of the custom 11T-LTQ-FT. Detection of methanethiol (MT; 9) via capture by 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB, Sigma-Aldrich) and HPLC analysis was performed as described previously (15) with the modification that all cell growth, feeding, and MT capture were performed under strict anaerobic conditions. Feedings were performed with 250 μm MTA and 10 μm [methyl-14C]MTA, prepared from S-adenosyl-[methyl-14C]methionine (PerkinElmer) as described (26); MT conjugated to DTNB was simultaneously detected at 372 nm optical absorption and by 14C radiometry on a Prominence HPLC (Shimadzu) with an in-line β-RAM 4 radio HPLC detector (IN/US Systems).
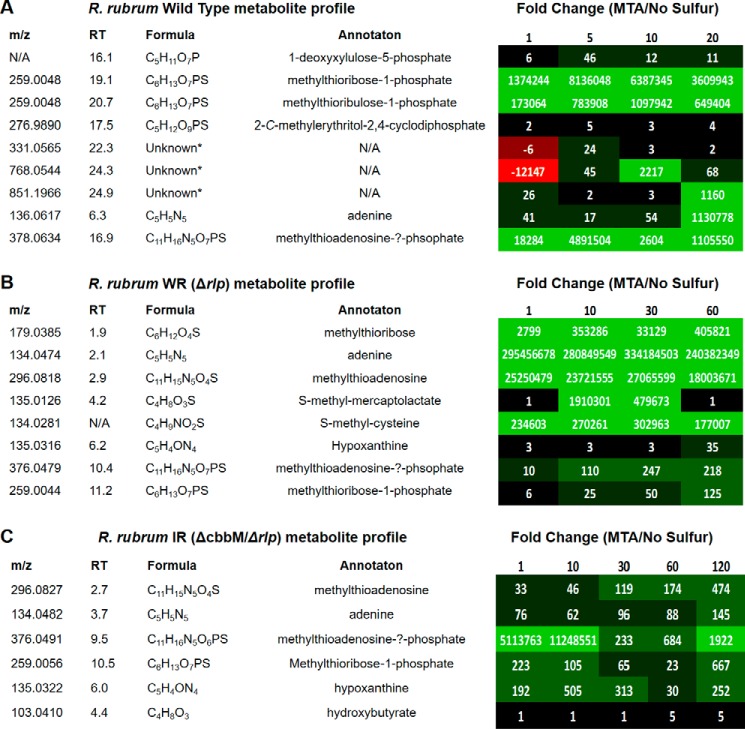
FIGURE 6.
Knock-out metabolomics analysis of R. rubrum strains. A–C, wild type (A), WR (ΔrlpA) (B), and IR (ΔcbbM/ΔrlpA) (C). The -fold change for each identified metabolite represents the relative metabolite concentration in cells fed with MTA versus no sulfur source under anaerobic conditions. Each column represents the time (min) after feeding. N/A, not applicable; RT, retention time.
Results
Requirement of Rubisco for Anaerobic MTA Metabolism
Previous studies of MTA metabolism in R. rubrum demonstrated that although RLP, which catalyzes the isomerization of MTRu-1P to MTXu-5P (27) (Fig. 1; H), is required for aerobic MTA metabolism, it is not required under anaerobic conditions (strain WR (ΔrlpA)) (Fig. 2, A and B) (14). However, deletion of the form II Rubisco (cbbM) gene (strains I19A (ΔcbbM) (17) and IR (ΔcbbM/ΔrlpA) (14)) resulted in the inability of the organism to allow significant photoheterotrophic growth on MTA as sole sulfur source, with or without the presence of a functional RLP (Fig. 2, B and D) (14). Moreover, Rubisco from R. rubrum does not appear to perform the MTRu-1P isomerization reaction catalyzed by B. subtilis RLP during MTA metabolism (15). These results suggest that Rubisco plays a functional role in anaerobic MTA metabolism in R. rubrum that differs from RLP and subsequent enzymes of the aerobic pathway (Fig. 1).
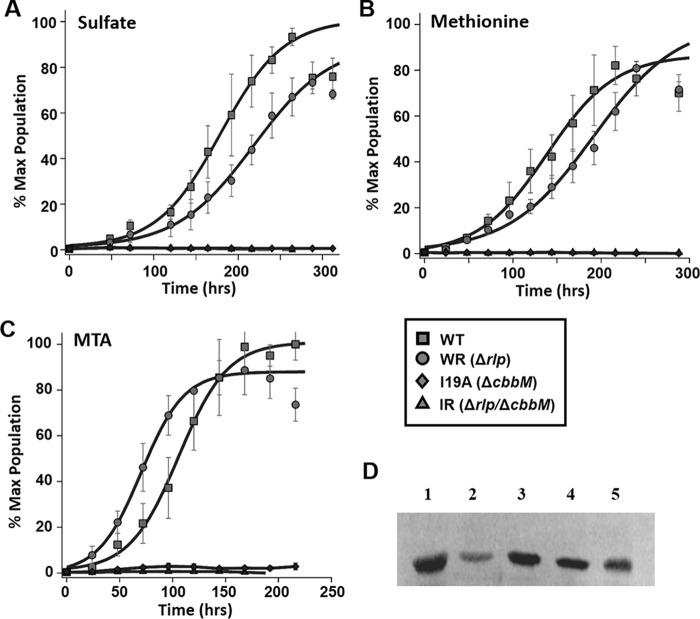
FIGURE 2.
Photoheterotrophic anaerobic growth in malate minimal medium of R. rubrum. A, wild type strain (white) and strain WR (ΔrlpA) (black) supplemented with 250 μm SO4−2 (squares), 250 μm MTA (circles), or without sulfur source (triangles). B, strain I19A (ΔcbbM) (white) and strain IR (ΔcbbM/ΔrlpA) (black) supplemented with 250 μm SO4−2 (squares), 250 μm MTA (circles), or without sulfur source (triangles). C, photoheterotrophic anaerobic growth in malate minima medium of R. rubrum wild type strain (white) and strain I19NifA (ΔcbbM,NifA-M173V) (black) supplemented with 250 μm SO4−2 (squares) and varying amounts of MTA at 1000 μm (circles), 200 μm (diamonds), 100 μm (triangles), and 50 μm (inverted triangle). D, photoheterotrophic anaerobic growth in malate minimal medium of R. rubrum strain IRNifA (ΔcbbM/ΔrlpA,NifA-M173V) supplemented with 250 μm SO4−2 (white squares), 250 μm MTA (white circles), or without sulfur source (white triangles) and of strain IRNifA grown under the same conditions with 250 μm MTA as sulfur source and complemented with pRPS-MCS3 plasmid containing the R. rubrum cbbM gene (black squares), R. palustris cbbM gene (black circles), or empty pRPS-MCS3 plasmid vector (black triangles). Microbial population data (means ± S.E. for n = 3 independent growth experiments) are plotted as the percentage of the maximum (% Max) OD660 nm of the positive control over growth time for each group. Data (excluding measurements beyond 75 h after onset of stationary phase) were fit to a sigmoidal logistic growth curve (solid lines) to estimate the average growth rate for each population (25) (Table 1).
To further address the requirement of Rubisco for anaerobic MTA metabolism, we determined the ability of the Rubisco deletion strain, I19NifA (ΔcbbM, nifA-M173V), to grow anaerobically on MTA as sole sulfur source as compared with wild type (Fig. 2C). Rubisco knock-out strains of R. rubrum are incapable of photoautotrophic growth (17, 28) and grow poorly under photoheterotrophic conditions using substrates such as malate as electron donor and carbon source and sulfate as the sulfur source (Fig. 2, Table 1). With such strains, the role of Rubisco and the CBB pathway in balancing the redox potential of the cell is taken over by other physiological processes, such as nitrogenase-dependent hydrogen evolution (28, 29). In the case of NifA-M173V, this mutation in the NifA transcriptional regulator of the nitrogen fixation genes (nif) leads to derepression of the nif operon, allowing Rubisco mutants to grow via dissipating excess reductant through the nitrogenase system (18, 28). This physiological adaptation provides a convenient means to monitor growth that is dependent on the sulfur source because the I19 strains have the means to dissipate reducing equivalents in the absence of a functional CBB pathway.
As with strain I19A, I19NifA Rubisco deletion strain was barely able to grow on MTA as sole sulfur source under anaerobic photoheterotrophic conditions over the range of MTA concentrations where growth of the wild type strain was observed (Fig. 2C). Growth of the wild type strain was not possible at MTA concentrations greater than 1 mm due to MTA toxicity, and less than 25–50 μm due to limiting MTA as the sulfur source (not shown). These results suggest that the inability of the R. rubrum I19NifA Rubisco deletion strain to grow anaerobically on MTA as sole sulfur source is not due to a redox imbalance caused by a nonfunctional CBB pathway.
Indications via knock-out mutations that Rubisco appeared to be specifically required for anaerobic MTA metabolism were further supported by complementation of the Rubisco/RLP double deletion strain, IRNifA (ΔcbbM/ΔrlpA,nifA-M173V) (Fig. 2D). Strains complemented with an empty pRPS-MCS3 plasmid (negative control) (19) could not restore anaerobic photoheterotrophic growth of these strains when MTA was used as sole sulfur source. However, expression in trans of a functional R. rubrum or R. palustris form II Rubisco (cbbM) inserted in the same pRPS-MCS3 plasmid restored MTA-dependent growth of strains IR (14) and IRNifA (Fig. 2D).
Rubisco Is Required for Both CO2 Fixation and MTA Metabolism
Prior studies did not consider the key question as to whether Rubisco might function concurrently as both a required carboxylase and a key catalyst for essential sulfur salvage reactions of the cell: e.g. simultaneously acting as a focal point for distinct carbon and sulfur metabolic pathways. To address this issue, the ability of R. rubrum to grow photoautotrophically when MTA was used as sole sulfur source and CO2 employed as sole carbon source was determined. Because photoautotrophic hydrogen-dependent and CO2-dependent growth absolutely requires a functional Rubisco for CO2 fixation to complete the CBB pathway (4), only strains that possessed a functional endogenous Rubisco (cbbM) gene were capable of MTA-dependent, CO2-dependent photoautotrophic growth (Fig. 3C), much like controls where either sulfate or methionine was provided as the source of sulfur (Fig. 3, A and B). Clearly, Rubisco was synthesized under both photoautotrophic and photoheterotrophic growth conditions irrespective of the sulfur source (Fig. 3D), with apparently higher amounts of protein synthesized (as seen in the immunoblots) when MTA was used as sulfur source under photoautotrophic growth conditions.
FIGURE 3.
R. rubrum photoautotrophic growth. A–C, photoautotrophic CO2-dependent growth of R. rubrum strains with 1 mm sulfate (A), 1 mm methionine (B), or 1 mm MTA (C) as sole sulfur source. Growth was performed in minimal medium bubbled with 1.5% CO2/98.5% H2. Microbial population data (means ± S.E. for n = 3) are plotted as the percentage of maximum (% Max) OD660 nm of positive control over growth time for each group and fit to a sigmoidal logistic growth curve (as described in the legend for Fig. 2) for WT strain (squares); strain WR (ΔrlpA) (circles); strain IR (ΔcbbM/ΔrlpA) (triangles); and strain I19A (ΔcbbM) (diamonds) (see Table 1 for growth rates). D, Western blots using antisera to R. rubrum Rubisco and extracts from various strains. Lane 1, WT R. rubrum grown photoautotrophically on 1 mm MTA as sole sulfur source; lane 2, WT strain grown photoheterotrophically on 1 mm MTA as sole sulfur source; lane 3, strain WR grown photoheterotrophically on 1 mm MTA as sole sulfur source; lane 4, strain WR grown photoautotrophically on 1 mm MTA as sole sulfur source; lane 5, strain WR grown photoautotrophically on 1 mm methionine as sole sulfur source.
Different Forms of Rubisco All Catalyze a Reaction Essential for Anaerobic MTA Metabolism in R. rubrum
Based on the complementation results with form II Rubiscos from R. rubrum and R. palustris (Fig. 2D), the question was raised as to whether other forms of bona fide Rubiscos were functionally capable of supporting MTA-dependent anaerobic phototrophic growth. Using the R. rubrum Rubisco/RLP double knock-out strain IR (ΔcbbM/ΔrlpA), complementation studies were employed with plasmid-borne exogenous Rubisco sequences added in trans to determine whether other diverse Rubisco genes might enable MTA-dependent growth. Plasmids containing both the form I and form II Rubisco genes (and endogenous promoter sequences) from R. sphaeroides (17) were able to support and complement MTA-dependent photoheterotrophic growth of the R. rubrum Rubisco/RLP deletion strain (Fig. 4, A and B).
FIGURE 4.
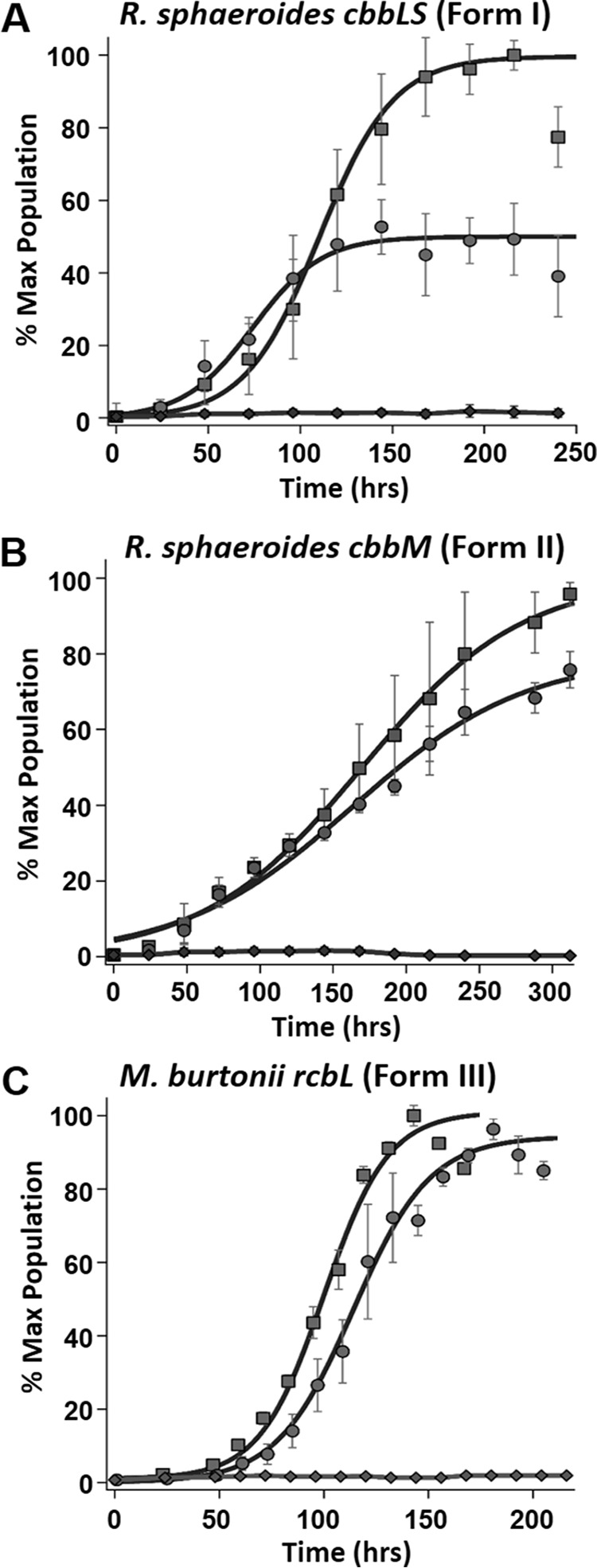
Complementation of Rubisco forms I, II, and III. A, complementation studies with R. rubrum strain IR (ΔcbbM/ΔrlpA) grown under anaerobic photoheterotrophic conditions in malate minimal medium and complemented with plasmid pJG336 from R. sphaeroides containing the form I Rubisco (cbbLS) genes with 1 mm sulfate (squares) and 1 mm MTA (circles) as sole sulfur source, as well as growth of strain IR on MTA lacking a Rubisco-containing plasmid (diamonds). B, photoheterotrophic growth of R. rubrum strain IR (ΔcbbM/ΔrlpA) complemented with R. sphaeroides plasmid pJG106 containing the form II Rubisco (cbbM) gene grown with 1 mm sulfate (squares) or 1 mm MTA (circles) as sole sulfur source, as well as growth of strain IR on MTA lacking a Rubisco-containing plasmid (diamonds). C, photoheterotrophic growth of R. rubrum strain IR (ΔcbbM/ΔrlpA) complemented with pRPS-MCS3 containing M. burtonii form III Rubisco (rbcL) gene with 1 mm sulfate (squares) or 1 mm MTA (circles) as sole source of sulfur and R. rubrum strain IR (ΔcbbM/ΔrlpA) grown on 1 mm MTA and complemented with empty vector (diamonds). Microbial population data (means ± S.E. for n = 3) are plotted as the percentage of maximum (% Max) OD660 nm of positive control over growth time for each group and fit to a sigmoidal logistic growth curve as described in the legend for Fig. 2(see Table 1 for growth rates).
These results strongly suggested that different forms of Rubisco from other organisms might also catalyze a reaction required for MTA-dependent growth in R. rubrum. Inasmuch as the R. sphaeroides plasmids contained sequences and unknown open reading frames in addition to Rubisco genes, the broad host complementation vector, pRPS-MCS3, was subsequently used to express only exogenous Rubisco genes in trans from the native R. rubrum promoter. It was determined that expression in trans of the evolutionarily distinct form I Rubisco (rbcLS) genes from the cyanobacterium, Synechococcus sp. strain PCC 6301 (20), rescued strain IR (ΔcbbM/ΔrlpA) for MTA-dependent photoheterotrophic growth (Fig. 5A). This is consistent with the observation that plasmids expressing Rubisco forms I and II from R. sphaeroides supported MTA-dependent photoheterotrophic growth of strain IR (ΔcbbM/ΔrlpA). Furthermore, the form III Rubisco (rbcL) gene from the methanogenic archaeon, M. burtonii (7, 30), also complemented strain IR with MTA as sole sulfur source (Fig. 4C). As with forms I and II, this archaeal form III Rubisco was presumably also capable of an essential sulfur salvage reaction in vivo to support MTA-dependent anaerobic photoheterotrophic growth. Clearly, all forms (I, II, and III) of bona fide Rubisco enabled anaerobic MTA-dependent growth in R. rubrum strain IR.
FIGURE 5.
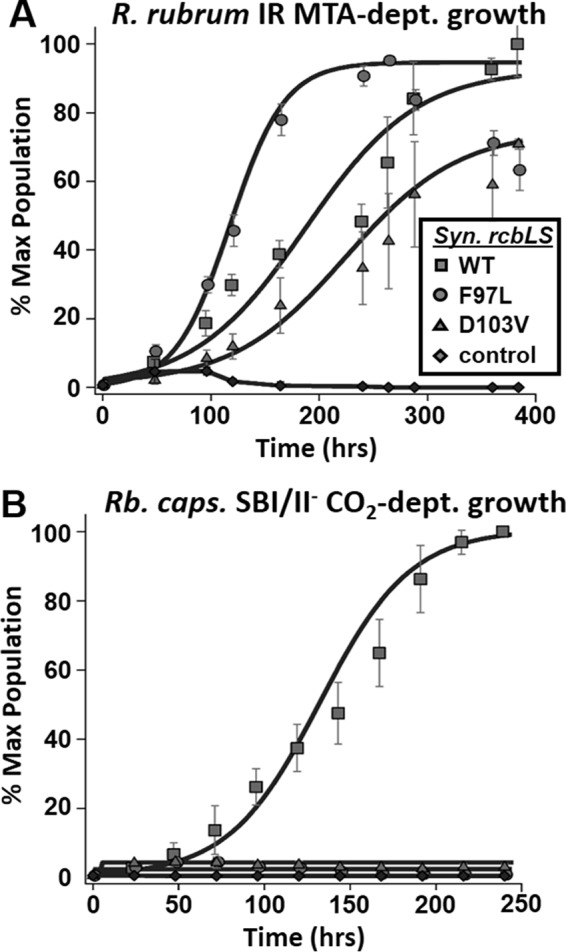
Complementation studies with anaerobic photoheterotrophically grown R. rubrum strain IR (ΔcbbM/ΔrlpA) in malate minimal medium with 1 mm MTA as sole sulfur source (A) and photoautotrophically grown R. capsulatus strain SBI/II− (ΔcbbLS/ΔcbbM) with CO2 as sole carbon source (B). In A and B, strains were complemented with pRPS-MCS3 containing Synechococcus sp. strain PCC 6301 wild type (squares) Rubisco genes (rcbLS), mutant F97L (circles), mutant D103V (triangles), or empty pRPS-MCS3 vector (diamonds). Microbial population data (means ± S.E. for n = 3) are plotted as the percentage of maximum (% Max) OD660 nm of positive control over growth time for each group and fit to a sigmoidal logistic growth curve as described in the legend for Fig. 2 (see Table 1 for growth rates). MTA-dept. growth, MTA-dependent growth; CO2-dept. growth, CO2-dependent growth.
Differential Effect of Active Site Residue Modifications on the CO2 Fixation and MTA Metabolism Functions of Rubisco
A number of Synechococcus Rubisco gene constructs containing mutations that alter the properties of this enzyme are available (19, 20, 31). For example, a previous study indicated that a change of residue Asp-103 to a Val on the large subunit (D103V mutant) negatively affected in vitro activity and resulted in the inability of the D103V enzyme to support CO2-dependent growth of a R. capsulatus Rubisco knock-out strain, strain SBI/II−(ΔcbbLS/ΔcbbM) (20). It is thought that this amino acid change leads to a negative growth phenotype by virtue of a disruption of interactions between large subunit dimers, instigating conformational changes that lead to diminished stability and/or functionality. In addition, a substitution at position Phe-97 to a Leu of the large subunit results in the formation of an enzyme with greatly diminished in vitro activity (20).
Constructs encoding the wild type and mutant cyanobacterial Rubiscos, D103V and F97L, were prepared in complementation vector pRPS-MCS3 and tested for their ability to complement R. capsulatus knock-out strain SBI/II− (ΔcbbLS/ΔcbbM) for anaerobic photoautotrophic CO2-dependent growth and R. rubrum strain IR (ΔcbbM/ΔrlpA) (Fig. 5A) for anaerobic photoheterotrophic MTA-dependent growth. Consistent with previous studies, these mutations in the large subunit of cyanobacterial Rubisco were not capable of supporting CO2-dependent growth of strain SBI/II− (Fig. 5B). However, we observed that both mutant enzymes enabled anaerobic photoheterotrophic growth of R. rubrum strain IR on MTA as sole sulfur source. Although these Rubisco mutant enzymes were compromised in their ability to catalyze CO2 fixation, they presumably were still able to catalyze an essential reaction required for sulfur salvage (MTA) metabolism. These results are consistent with the interpretation that at least some amino acid residues that are required for carboxylase function of Rubisco are not essential for anaerobic MTA-dependent growth.
Rubisco in R. rubrum May Link Anaerobic MTA Metabolism to S-methyl-cysteine Production
To eventually identify the reaction catalyzed by Rubisco to support anaerobic MTA-dependent growth, we employed the recently developed knock-out metabolomics approach previously used to determine the complete aerobic MTA metabolic pathway and novel isoprenoid shunt in R. rubrum (13). Comparison of metabolite profiles from wild type and knock-out strains fed with MTA versus no sulfur source enables identification of specific metabolites that are potentially metabolized by enzymes encoded by the gene(s) in question.
Metabolomics profiling of the R. rubrum WT strain fed with MTA under anaerobic, photoheterotrophic conditions showed the presence of most known aerobic MTA metabolites (i.e. MTR-1P, MTRu-1P, DXP, cMEPP) (Fig. 6A). Furthermore, MT generation, which is the last step before methionine synthesis in the aerobic pathway (Fig. 1; I, 9), was confirmed during anaerobic MTA metabolism by HPLC analysis of MT conjugation to DTNB (Fig. 7). Given that knock-out strain growth analysis showed that R. rubrum RLP was not required to support anaerobic MTA-dependent growth, whereas Rubisco was (Fig. 2, A and B), it was surprising that the only observed metabolites up-regulated in the wild type strain fed with MTA corresponded to the known aerobic pathway in which the RLP is required (Fig. 1) (13). This suggested that a pathway similar to the R. rubrum aerobic pathway in which the RLP participates functions under anaerobic conditions as well. However, the functions of Rubisco and RLP under anaerobic growth were not resolved.
FIGURE 7.
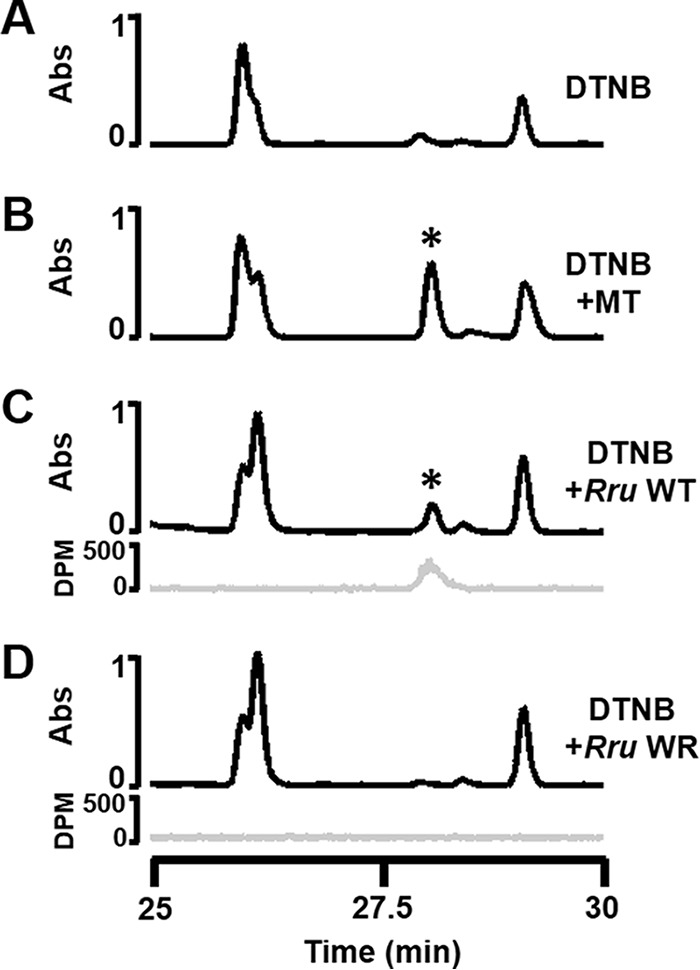
HPLC detection of methanethiol generated. A, unconjugated DTNB detected at 372 nm (black line). B, MT standard (Spex Certi-Prep) conjugated to DTNB detected at 372 nm (black line), * indicates peak corresponding to DTNB-methanethiol adduct. C and D, DTNB incubated with gaseous headspace generated during [methyl-14C]MTA feeding of R. rubrum WT strain (C) and R. rubrum strain WR (ΔrlpA) assayed by HPLC with detection at 372 nm (black line) and by 14C radio-HPLC (gray line) (D).
To separate the RLP and Rubisco components, metabolomics profiling was performed on the R. rubrum RLP deletion strain, WR (ΔrlpA), fed with MTA (Fig. 6B) under anaerobic conditions. Consistent with the lack of a functional RLP, no metabolites downstream of the RLP reaction in the known aerobic pathway were observed, nor was any MT generation detected under anaerobic conditions (Fig. 7). Furthermore, two previously unobserved metabolites with chemical formula C4H9NO2S and C4H8O3S, suggestive of S-methyl-cysteine and S-methyl-mercaptolactate (Fig. 1, 14, 15), were observed during anaerobic feedings with MTA. To determine the dependence of these metabolites on Rubisco activity, we measured the metabolomics profile of the Rubisco/RLP double deletion strain, IR (ΔcbbM/ΔrlpA), fed with MTA (Fig. 6C). In the absence of a functional Rubisco, both S-methyl-mercaptolactate and S-methyl-cysteine were no longer observed. Although it is uncertain whether Rubisco acts directly or indirectly to form these metabolites, these results suggest that under anaerobic conditions, sulfur is salvaged from MTA presumably through S-methyl-mercaptolactate and S-methyl-cysteine via a pathway that explicitly requires Rubisco.
Discussion
It is apparent that Rubisco involvement in sulfur (MTA) metabolism in R. rubrum is limited to anaerobic phototrophic growth, yet the same enzyme plays a key and concurrent role in essential carbon fixation. These discrete functions may be partially separated, as certain mutant Rubisco proteins show a differential effect on either carboxylation or sulfur salvage functions, exemplified by the ability of the D103V and F97L mutant cyanobacterial form I enzymes to support MTA-dependent growth but not CO2-dependent growth in the proper indicator host strains: e.g. R. rubrum strain IR or R. capsulatus strain SBI/II−, respectively. In addition, in R. rubrum, prior studies suggest the possibility that poor photoheterotrophic growth of Rubisco mutants may be due in part to an inhibitory toxic buildup of RuBP (32). However, in order for such strains to grow photoheterotrophically using MTA as sole sulfur source, a Rubisco gene functional for MTA metabolism is required and must be added in trans (14). Because the Rubisco CO2 fixation function is compromised in the aforementioned mutant enzymes, it is apparent that Rubisco CO2 fixation is not simply removing inhibitory RuBP (32) or allowing the CBB pathway to alleviate some cellular redox imbalance (18, 28, 29, 32) to support MTA-dependent anaerobic growth. Rather, the ability of the CO2 fixation-compromised mutant enzymes to complement R. rubrum strain IR (ΔcbbM/ΔrlpA) for MTA-dependent photoheterotrophic growth suggests that Rubisco is providing some key role in MTA metabolism. Further, these results also suggest that the active site of Rubisco may be somewhat different for the carboxylation and key sulfur salvage reactions presumably catalyzed by the enzyme. Moreover, it appears that the active site of all three distinct forms (I, II, and III) of bona fide Rubisco has been similarly modified by evolution in one important respect, as all three forms appear to be utilized for both the carboxylation and MTA metabolism functions. These results further suggest that the two functions of Rubisco have distinct and important physiological relevance in many, if not all organisms that contain this enzyme, as clearly exemplified by strains of R. rubrum where growth is impossible unless both carboxylation and MTA metabolic reactions take place.
Finally, there remain many interesting questions that need to be addressed. For one, it is not clear what the precise role of the Rubisco-catalyzed MTA metabolism reaction might be in vivo because there do not appear to be recognizable genes that encode proteins for subsequent anaerobic MTA metabolism beyond the canonical MTA phosphorylase (Rru_0361) and MTR-1P isomerase (Rru_0360) genes (Fig. 1; F, G). Inasmuch as Rubisco is notorious for catalyzing several side reactions (see Ref. 33 and references therein), could it be possible that one of these or some unknown side reaction plays an important role in anaerobic MTA metabolism?
The finding that compounds consistent with S-methyl-mercaptolactate and S-methyl-cysteine were synthesized when a functional Rubisco was present provides several insights. The first consideration is the potential mechanism by which these compounds are synthesized. In Arabidopsis sp., O-acetylserine (thiol) lyase catalyzes the condensation of generated methanethiol with O-acetylserine to form S-methyl-cysteine (34). However, given the absence of methanethiol production when the R. rubrum RLP is inactivated, a methanethiol intermediate is unlikely. Alternatively, this suggests either a C–C lyase mechanism to produce S-methyl-mercaptolactate or a sulfurtransferase mechanism (35) by which a putative sulfurtransferase catalyzes the transfer of the methylthiol group to pyruvate to form S-methyl-mercaptolactate. Further work is required to determine whether Rubisco is directly or indirectly involved in S-methyl-cysteine and S-methyl-mercaptolactate metabolism.
Secondly, it will be important to determine how these metabolites are utilized for regenerating usable sulfur. Recently, S-methyl-cysteine metabolic pathways paralogous to cysteine metabolism were discovered in B. subtilis, in which S-methyl-cysteine was metabolized to methionine without the requirement of sulfur oxygenation (36). However, perusal of the genome indicates that this same pathway appears to be lacking in R. rubrum. At this juncture, the fate of S-methyl-cysteine and S-methyl-mercaptolactate in R. rubrum is largely unknown, but nevertheless these compounds are metabolized in the Rubisco-dependent anaerobic sulfur (MTA) salvage pathway.
Clearly, it will be of interest to identify the reaction catalyzed by Rubisco for anaerobic phototrophic MTA metabolism, elucidate the presumptive novel anaerobic MTA metabolic pathway, and eventually determine how these diverse sulfur salvage pathways, involving both Rubisco and RLP, are differentially controlled in the cell. The concurrent involvement of Rubisco as an essential and physiologically significant catalyst for both central carbon and sulfur metabolism suggests the evolution of a heretofore unappreciated and major functional versatility for this protein.
Author Contributions
S. D., J. A. N., J. S., and B. S. E. conducted the experiments and performed the statistical analyses and data processing of all growth and metabolomics data. F. R. T. designed the experiments and supervised the research. All authors discussed the research and contributed to writing the manuscript.
Acknowledgments
We thank Dr. Kevin Sowers of the University of Maryland, Center of Marine Biotechnology for providing M. burtonii genomic DNA and Dr. Brian Witte for constructing plasmid pRPS-MBR. We thank Dr. Sriram Satagopan and Dr. Vanessa Varaljay for many useful conversations. This work was also supported by the National Science Foundation under Grant DBI-0922879 for acquisition of the LTQ-Velos Pro Orbitrap LC-MS/MS (to B. S. E.).
This work was supported by National Institutes of Health Grant GM095742 (to F. R. T.), and by a Ruth L. Kirschstein National Research Service Award (F32GM109547) from the NIGMS (to J. A. N.) The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as a Paper of the Week.
- CBB
- Calvin-Benson-Bassham
- cMEPP
- 2-C-methylerythritol-2,4-cyclodiphosphate
- DK-MTP-1P
- 2,3-diketo-5-methylthiopentyl-1-phosphate
- DTNB
- 5,5′-dithiobis-(2-nitrobenzoic acid)
- DXP
- 1-deoxyxylulose-5-phosphate
- HK-MTPene-1P
- 2-hydroxy-3-keto-5-methylthiopent(1)ene-1-phosphate
- MT
- methanethiol
- MTA
- 5-methylthioadenosine
- MTR-1P
- 5-methylthioribose-1-phosphate
- MTRu-1P
- 5-methylthioribulose-1-phosphate
- MTXu-5P
- 1-methylthioxylulose-5-phosphate
- RLP
- Rubisco-Like protein
- Rubisco
- ribulose-1,5-bisphosphate carboxylase/oxygenase
- RuBP
- ribulose-1,5-bisphosphate.
References
- 1. Cleland W. W., Andrews T. J., Gutteridge S., Hartman F. C., and Lorimer G. H. (1998) Mechanism of RubisCO: the carbamate as general base. Chem. Rev. 98, 549–562 [DOI] [PubMed] [Google Scholar]
- 2. Spreitzer R. J., and Salvucci M. E. (2002) Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 53, 449–475 [DOI] [PubMed] [Google Scholar]
- 3. Hauser T., Popilka L., Hartl F. U., and Hayer-Hartl M. (2015) Role of auxiliary proteins in Rubisco biogenesis and function. Nature Plants 1, 15065. [DOI] [PubMed] [Google Scholar]
- 4. Tabita F. R. (1999) Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth. Res. 60, 1–28 [Google Scholar]
- 5. Hanson T. E., and Tabita F. R. (2001) A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 98, 4397–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tabita F. R., Hanson T. E., Li H., Satagopan S., Singh J., and Chan S. (2007) Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol. Mol. Biol. Rev. 71, 576–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tabita F. R., Hanson T. E., Satagopan S., Witte B. H., and Kreel N. E. (2008) Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Phil. Trans. R. Soc. Lond. B Biol. Sci. 363, 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H., Sawaya M. R., Tabita F. R., and Eisenberg D. (2005) Crystal structure of a novel RuBisCO-like protein from the green sulfur bacterium Chlorobium tepidum. Structure 13, 779–789 [DOI] [PubMed] [Google Scholar]
- 9. Hanson T. E., and Tabita F. R. (2003) Insights into the stress response and sulfur metabolism revealed by proteome analysis of a Chlorobium tepidum mutant lacking the RubisCO-like protein. Photosynth. Res. 78, 231–248 [DOI] [PubMed] [Google Scholar]
- 10. Ashida H., Saito Y., Kojima C., Kobayashi K., Ogasawara N., and Yokota A. (2003) A functional link between Rubisco-like protein of Bacillus and photosynthetic Rubisco. Science 302, 286–290 [DOI] [PubMed] [Google Scholar]
- 11. Carré-Mlouka A., Méjean A., Quillardet P., Ashida H., Saito Y., Yokota A., Callebaut I., Sekowska A., Dittmann E., Bouchier C., and de Marsac N. T.(2006) A new RubisCO-like protein coexists with a photosynthetic Rubisco in the planktonic cyanobacteria Microcystis. J. Biol. Chem. 281, 24462–24471 [DOI] [PubMed] [Google Scholar]
- 12. Imker H. J., Fedorov A. A., Fedorov E. V., Almo S. C., and Gerlt J. A. (2007) Mechanistic diversity in the RuBisCO superfamily: the “enolase” in the methionine salvage pathway in Geobacillus kaustophilus. Biochemistry 46, 4077–4089 [DOI] [PubMed] [Google Scholar]
- 13. Erb T. J., Evans B. S., Cho K., Warlick B. P., Sriram J., Wood B. M., Imker H. J., Sweedler J. V., Tabita F. R., and Gerlt J. A. (2012) A RubisCO-like protein links SAM metabolism with isoprenoid biosynthesis. Nat. Chem. Biol. 8, 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh J., and Tabita F. R. (2010) Role of RubisCO and the RubisCO-like protein in 5-methyladenosine metabolism in the nonsulfur bacterium Rhodospirillum rubrum. J. Bacteriol. 192, 1324–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Warlick B. P., Imker H. J., Sriram J., Tabita F. R., and Gerlt J. A. (2012) Mechanistic Diversity in the RubisCO Superfamily: The RubisCO from Rhodospirillum rubrum is not promiscuous for reactions catalyzed by RubisCO-like proteins (RLPs). Biochemistry 51, 9470–9479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Munk A. C., Copeland A., Lucas S., Lapidus A., Del Rio T. G., Barry K., Detter J. C., Hammon N., Israni S., Pitluck S., Brettin T., Bruce D., Han C., Tapia R., Gilna P., Schmutz J., Larimer F., Land M., Kyrpides N. C., Mavromatis K., Richardson P., Rohde M., Göker M., Klenk H. P., Zhang Y., Roberts G. P., Reslewic S., and Schwartz D. C. (2011) Complete genome sequence of Rhodospirillum rubrum type strain (S1). Stand. Genomic Sci. 4, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Falcone D. L., and Tabita F. R. (1993) Complementation analysis and regulation of CO2 fixation gene expression in a ribulose-1,5-bisphosphate carboxylase-oxygenase deletion strain of Rhodospirillum rubrum. J. Bacteriol. 175, 5066–5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang D., Zhang Y., Welch E., Li J., and Roberts G. P. (2010) Elimination of RubisCO alters the regulation of nitrogenase activity and increases hydrogen production in Rhodospirillum rubrum. Int. J. Hydrogen Energy 35, 7377–7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith S. A., and Tabita F. R. (2003) Positive and negative selection of mutant forms of prokaryotic (cyanobacterial) ribulose-1, 5-bisphosphate carboxylase/oxygenase. J. Mol. Biol. 331, 557–569 [DOI] [PubMed] [Google Scholar]
- 20. Satagopan S., Scott S. S., Smith T. G., and Tabita F. R. (2009) A RubisCO mutant that confers growth under a normally “inhibitory” oxygen concentration. Biochemistry 48, 9076–9083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Witte B. H. (2012) Taming the Wild RubisCO: Explorations in Functional Metagenomics. Ph.D. thesis, The Ohio State University [Google Scholar]
- 22. Satagopan S., Chan S., Perry L. J., and Tabita F. R. (2014) Structure-function studies with the unique hexameric form II ribulose-1,5-bisphosphate carboxylase/oxygenase (RubsiCO) from Rhodospeudomonas plaustris. J. Biol. Chem. 289, 21433–21450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gibson J. L., and Tabita F. R. (1986) Isolation of the Rhodopseudomonas sphaeroides form I ribulose 1,5-bisphosphate carboxylase/oxygenase large and small subunit genes and expression of the active hexadecameric enzyme in Escherichia coli. Gene 44, 271–278 [DOI] [PubMed] [Google Scholar]
- 24. Ormerod J. G., Ormerod K. S., and Gest H. (1961) Light-dependent utilization of organic compounds and photoproduction of molecular hydrogen by photosynthetic bacteria; relationships with nitrogen metabolism. Arch. Biochem. Biophys. 94, 449–463 [DOI] [PubMed] [Google Scholar]
- 25. Peleg M., and Corradini M. G. (2011) Microbial growth curves: what the models tell us and what they cannot. Crit. Rev. Food Sci. Nutr. 51, 917–945 [DOI] [PubMed] [Google Scholar]
- 26. Schlenk F., and Ehninger J. (1964) Observation of 5′-methylthioadenosine. Arch. Biochem. Biophys. 106, 95–100 [DOI] [PubMed] [Google Scholar]
- 27. Imker H. J., Singh J., Warlick B. P., Tabita F. R., and Gerlt J. A. (2008) Mechanistic diversity in the RubisCO superfamily: a novel isomerization reaction catalyzed by the Rubisco-like protein from Rhodospirillum rubrum. Biochemistry 47, 11171–11173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Joshi H. M., and Tabita F. R. (1996) A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc. Natl. Acad. Sci. U.S.A. 93, 14515–14520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McKinlay J. B., and Harwood C. S. (2010) Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 11669–11675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alonso H., Blayney M. J., Beck J. L., and Whitney S. M. (2009) Substrate-induced assembly of Methanococcoides burtonii d-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers. J. Biol. Chem. 284, 33876–33882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith S. A., and Tabita F. R. (2004) Glycine 176 affects catalytic properties and stability of the Synechococcus sp. Strain PCC 6301 ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Biol. Chem. 279, 25632–25637 [DOI] [PubMed] [Google Scholar]
- 32. Wang D., Zhang Y., Pohlmann E. L., Li J., and Roberts G. P. (2011) The poor growth of Rhodospirillum rubrum mutants lacking RubisCO is due to the accumulation of ribulose-1,5-bisphosphate. J. Bacteriol. 193, 3293–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim K., and Portis A. R. Jr. (2004) Oxygen-dependent H2O2 production by RubisCO. FEBS Lett. 571, 124–128 [DOI] [PubMed] [Google Scholar]
- 34. Rébeillé F., Jabrin S., Bligny R., Loizeau K., Gambonnet B., Van Wilder V., Douce R., and Ravanel S. (2006) Methionine catabolism in Arabidopsis cells is initiated by a γ-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc. Natl. Acad. Sci. U.S.A. 103, 15687–15692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cipollone R., Ascenzi P., and Visca P. (2007) Common themes and variations in the rhodanese superfamily. IUBMB Life 59, 51–59 [DOI] [PubMed] [Google Scholar]
- 36. Chan C. M., Danchin A., Marlière P., and Sekowska A. (2014) Paralogous metabolism: S-alkyl-cysteine degradation in Bacillus subtilis. Environ. Microbiol. 16, 101–117 [DOI] [PubMed] [Google Scholar]