FIGURE 4.
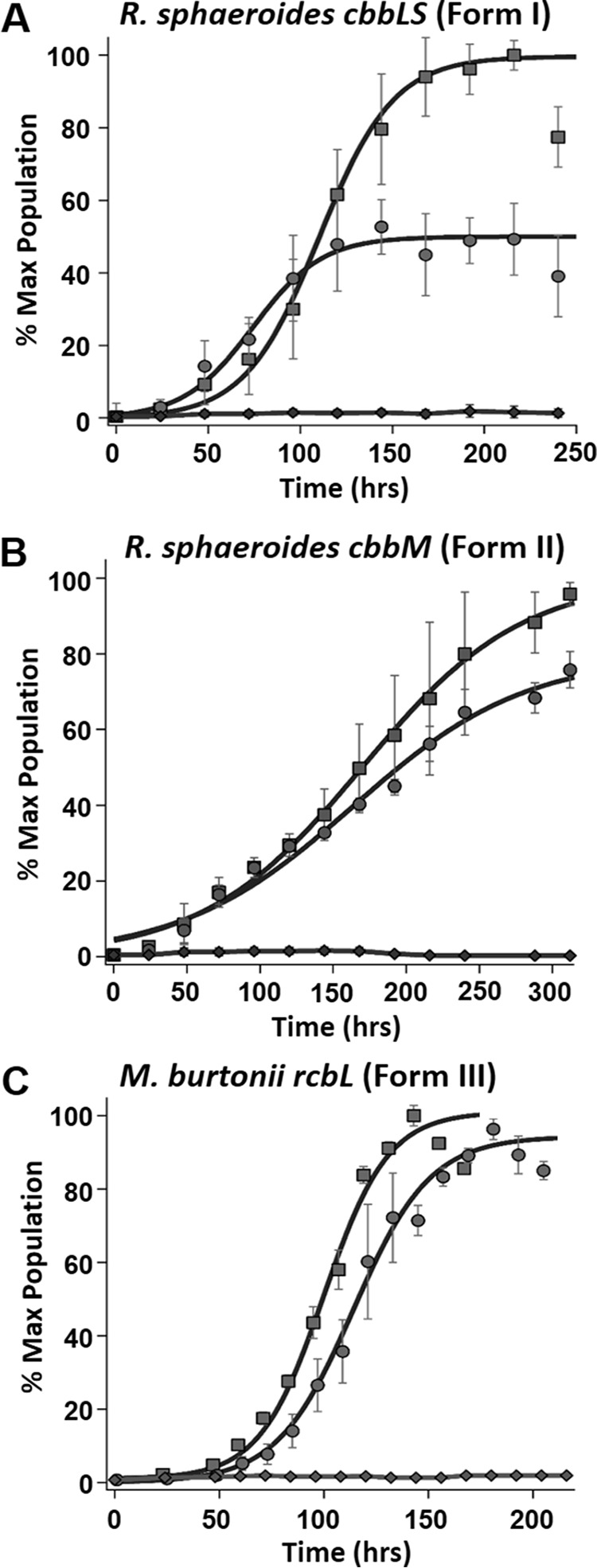
Complementation of Rubisco forms I, II, and III. A, complementation studies with R. rubrum strain IR (ΔcbbM/ΔrlpA) grown under anaerobic photoheterotrophic conditions in malate minimal medium and complemented with plasmid pJG336 from R. sphaeroides containing the form I Rubisco (cbbLS) genes with 1 mm sulfate (squares) and 1 mm MTA (circles) as sole sulfur source, as well as growth of strain IR on MTA lacking a Rubisco-containing plasmid (diamonds). B, photoheterotrophic growth of R. rubrum strain IR (ΔcbbM/ΔrlpA) complemented with R. sphaeroides plasmid pJG106 containing the form II Rubisco (cbbM) gene grown with 1 mm sulfate (squares) or 1 mm MTA (circles) as sole sulfur source, as well as growth of strain IR on MTA lacking a Rubisco-containing plasmid (diamonds). C, photoheterotrophic growth of R. rubrum strain IR (ΔcbbM/ΔrlpA) complemented with pRPS-MCS3 containing M. burtonii form III Rubisco (rbcL) gene with 1 mm sulfate (squares) or 1 mm MTA (circles) as sole source of sulfur and R. rubrum strain IR (ΔcbbM/ΔrlpA) grown on 1 mm MTA and complemented with empty vector (diamonds). Microbial population data (means ± S.E. for n = 3) are plotted as the percentage of maximum (% Max) OD660 nm of positive control over growth time for each group and fit to a sigmoidal logistic growth curve as described in the legend for Fig. 2(see Table 1 for growth rates).
