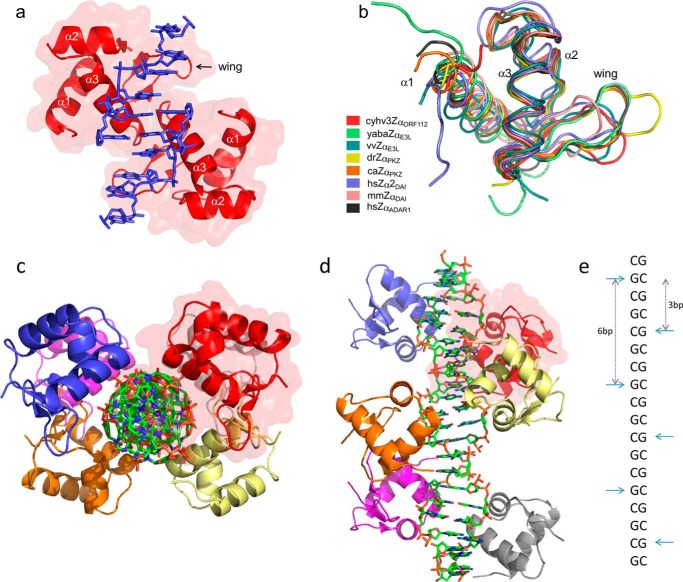
FIGURE 1.
Overall structure of Cyprinid herpesvirus 3 Zα domain in complex with Z-DNA and its fold similarity to other members of Zα family. a, representation of the asymmetric unit of CyHV-3 ORF112-Zα: two protein domains (red with semi-transparent surface) and two Z-DNA strands (in blue). b, structural alignments (Cα trace) of Zα family members: Cyprinid herpesvirus 3 ORF112-Zα (cyhv3ZαORF112, PDB code 4WCG), Yaba-like E3L-Zα (yabaZαE3L, PDB code 1SFU), vaccinia virus E3L-Zα (vvZαE3L, PDB code 1OYI), D. rerio PKZ-Zα (drZαPKZ, PDB code 4LB5), C. auratus PKZ-Zα (caZαPKZ, PDB code 4KMF), Homo sapiens DAI-Zα2 (hsZα2DAI, PDB code 3EYI), Mus musculus DAI-Zα (mmZαDAI, PDB code 1J75), and Homo sapiens ADAR1-Zα (hsZαADAR1, PDB code 1QBJ). c and d, representation of reconstructed biological assembly of T(CG)9 oligonucleotide. The view is perpendicular to (c) or along (d) the DNA axis. DNA duplex is represented as a stick model. Proteins are depicted as the ribbons; six monomers decorate T(CG)9 oligonucleotide. Red transparent surface marking one of the monomers serves as a reference. e, schematic representation of the arrangement of the ORF112-Zα domains along the Z-DNA helix. The CH-π interaction between the guanosines in the syn conformation and Tyr-257 is used as the reference and is indicated by horizontal cyan arrows. Vertical arrows indicate binding site spacing (number of base pairs) between adjacent monomers.