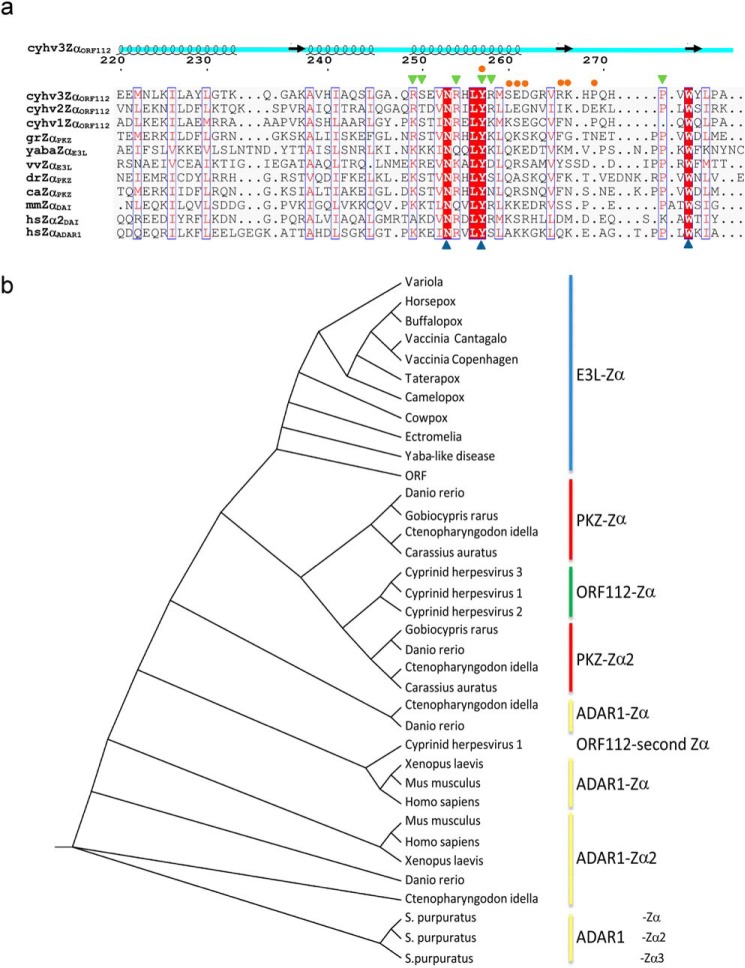
FIGURE 4.
ORF112-Zα phylogenetic analysis. a, structure-aided alignment of CyHV-3 ORF112-Zα with other Zα domains (for PDB codes see Fig. 1). Three sequences without structural information are included: Cyprinid herpesvirus 1 (cyhv1ZαORF112), Cyprinid herpesvirus 2 (cyhv2ZαORF112), and G. rarus PKZ-Zα (grZαPKZ). On the top of the alignment, the schematic representation of ORF112-Zα (cyhv3ZαORF112) secondary structure is drawn. Blue triangles below the alignment mark the triad of critical Z-DNA/Z-RNA binding residues: Asn, Tyr, and Trp. Blue boxes mark positions with conservation higher than 50%, and red shading highlights absolute conservation in this alignment. Residues involved in protein·DNA (green triangles) and protein-protein interactions (orange circles) are indicated above the alignment. b, cladogram generated based on curated (gaps removed) Muscle alignment with PhyML using Jones-Taylor-Thornton substitution model with the Shimodaira-Hasegawa approximate likelihood ratio test.