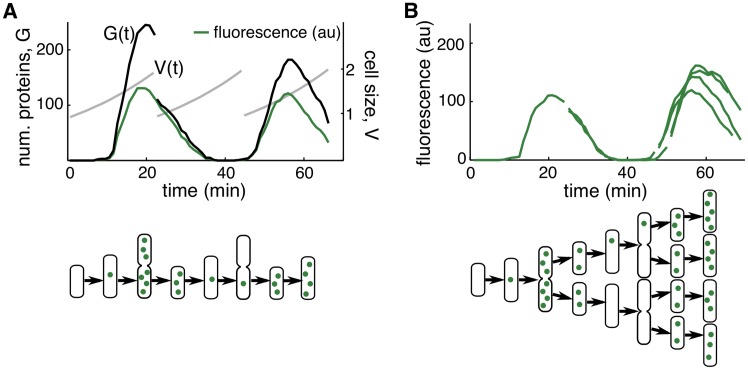
Fig 4. Incorporating the cell cycle into stochastic simulations.
A. In a single cell simulation, a cell’s dynamics are modeled until it divides. After division the cell volume (V(t), gray) is halved and proteins (G(t), black) are partitioned binomially. Only one of the daughter cells is used in the remainder of the simulation. The concentration of G is G(t)/V(t) which results in a single cell trajectory (green). B. In a lineage simulation the volume of a cell and the number of proteins upon division are treated similarly to a single cell simulation. However, instead of throwing one daughter cell away at each division, the dynamics of both daughter cells are modeled and a lineage trajectory (green) is obtained.