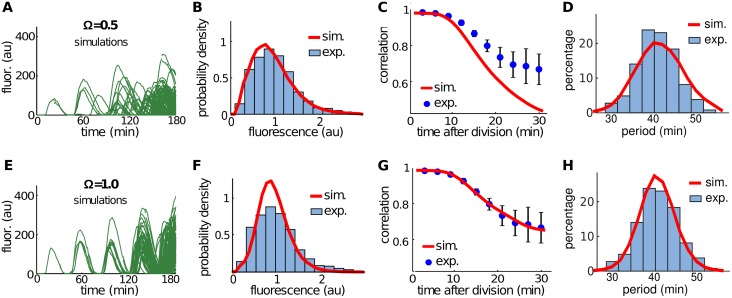
Fig 5. Lineage simulations in the absence of extrinsic noise.
We chose the scaling parameter Ω to fit the amplitude variability (A-D, Ω = 0.5) or correlation between sister cells (E-H, Ω = 1.0) obtained from experimental data. A, E. Typical lineage simulations. B, F. Distribution of the oscillation amplitudes for experimental data (blue histograms) and from simulations (red curves), with amplitude rescaled by the mean. The coefficient of variation of the amplitude equals 0.47 for experiments, 0.47 for panel B, and 0.35 for panel F. C,G. Correlation in fluorescence intensity between sister cells after cell division for experimental data (blue circles, mean ± s.d.) and simulations (red curves). D,H. Distribution of the period for experimental data (blue histograms) and from simulations (red curves). The coefficient of variation of the period equals 0.11 for experiments, 0.14 for panel B, and 0.10 for panel F.