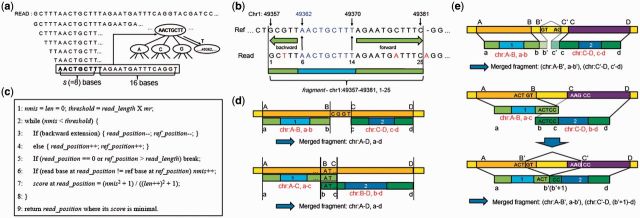
Fig. 2.
Read alignment. (a) Sliding windows are applied to a read with a window size s + 16 and step size 1. Genomic positions stored in the index are retrieved using the windowed sequence as illustrated. (b) An example to illustrate the extension process for a specific window with sequence ‘AACTGCTTT’ (blue) that was aligned to position 49362. The extension was carried out in both backward and forward directions. Mismatches and indels are labeled in red. (c) Pseudo code for the extension process in (b). (d) Merging of two fragments whose distance is less than the allowed number of insertions or deletions. Individual fragments are labeled in red. (e) Merging of two fragments whose distance is greater than the allowed number of insertions or deletions, which are examined as candidate spliced junction reads. Splice sites are determined using the GT-AG or GC-AG rules