Summary
Group 2 innate lymphoid cells (ILC2s) produce a significant amount of interleukin‐5 (IL‐5), which supports eosinophil responses in various tissues; they also produce IL‐13, which induces mucus production and contributes to tissue repair or fibrosis. The ILC2s are activated by alarmins, such as IL‐33 released from epithelia, macrophages and natural killer T (NKT) cells in response to infection and allergen exposure, leading to epithelial injury. We examined gene expression in lung ILC2s and found that ILC2s expressed Ifngr1, the receptor for interferon‐γ (IFN‐γ). Interferon‐γ severely inhibited IL‐5 and IL‐13 production by lung and kidney ILC2s. To evaluate the effects in vivo, we used α‐galactosylceramide (α‐GalCer) to induce NKT cells to produce IL‐33 and IFN‐γ. Intraperitoneal injection of α‐GalCer in mice induced NKT cell activation resulting in IL‐5 and IL‐13 production by ILC2s. Administration of anti‐IFN‐γ together with α‐GalCer significantly enhanced the production of IL‐5 and IL‐13 by ILC2s in lung and kidney. Conversely, cytokine production from ILC2s was markedly suppressed after injection of exogenous IL‐33 in Il33 −/− mice pre‐treated with α‐GalCer. Hence, IFN‐γ induced or already present in tissues can impact downstream pleiotropic functions mediated by ILC2s, such as inflammation and tissue repair.
Keywords: group 2 innate lymphoid cells, interferon‐γ, interleukin‐5, interleukin‐13, T helper type 2 cytokines
Abbreviations
- IFN‐γ
interferon‐γ
- ILC2s
group 2 innate lymphoid cells
- IL
interleukin
- NK
natural killer
- TSLP
thymic stromal lymphopoietin
- α‐GalCer
α‐galactosylceramide
Introduction
Type 2 immune responses are elicited by parasites, allergens, and inhaled proteases.1, 2 The type 2 cytokines interleukin‐4 (IL‐4), IL‐5, IL‐9 and IL‐13 are primarily secreted by adaptive CD4+ T helper type 2 (Th2) cells, but also by the recently identified group 2 innate lymphoid cells (ILC2s).3, 4, 5 ILC2s are present in mesenteric fat‐associated lymphoid clusters and non‐lymphoid tissues, including the lung, liver, skin, intestine and visceral adipose tissue of mice6, 7, 8, 9 and humans.10, 11
The ILC2s contribute to type 2 immune responses in response to allergic inflammation and tissue repair12, 13, 14 by producing high amounts of IL‐5 and IL‐13. Interleukin‐5 is involved in many aspects of eosinophil development and accumulation in various tissues, and supports the growth and differentiation of activated B cells, including mucosal IgA production and the maintenance of B‐1 cells in mice.15 Local production of IL‐5 in lung is associated with eosinophil accumulation and the exacerbation of inflammatory responses,14, 16 whereas eosinophils can also promote the regeneration of damaged tissues.17 We have reported that IL‐5‐producing ILC2s are present in lung and that exposure to IL‐33 induces IL‐5 production by ILC2s, resulting in the accumulation of eosinophils in lung and the suppression of cancer metastasis.18 The ILC2s are also involved in promoting tissue repair, remodelling and fibrotic responses through the production of IL‐13, which promotes goblet cell mucus secretion and smooth muscle contraction,19 and amphiregulin, an epidermal growth factor (EGF)‐like molecule that promotes tissue integrity following infection and inflammation.20
Group 2 innate lymphoid cells are activated by stimulation with epithelial‐derived cytokines, including IL‐33, IL‐25 and thymic stromal lymphopoietin (TSLP).21 During viral infections, activated natural killer T (NKT) cells and alveolar macrophages also produce IL‐33, which stimulates ILC2s, leading to further cytokine production.22, 23 In addition, ILC2s are activated by IL‐9,24 basophil‐derived IL‐4,25 prostaglandin D2,26 and leukotrienes.27 Signalling by inducible T‐cell co‐stimulator (ICOS) and its ligand can promote cytokine production and ILC2 survival through signal transducer and activator of transcription 5 (STAT5) signalling.28 However, suppression of ILC2 function has not been carefully examined.
Naive CD4+ T (Th0) cells differentiate into Th1 or Th2 cells,29 and type 1 and type 2 cytokines cross‐regulate Th1 and Th2 development and expansion.30, 31, 32 Interferon‐γ (IFN‐γ), a type 1 cytokine secreted by Th1 cells, CD8+ effector T cells, NK cells, NKT cells and other cells in response to acute inflammation or viral infection,33, 34, 35, 36 is a key regulator that initiates type 1 immune responses while suppressing type 2 immunity.37, 38 However, the ability of IFN‐γ to antagonize ILC2 function has not been properly evaluated.
In this report we show that ILC2s express Ifngr1, the receptor for IFN‐γ, and that cytokine production by ILC2s is down‐regulated by IFN‐γ. Production of IL‐5 and IL‐13 by lung and kidney ILC2s was inhibited by IFN‐γ derived from activated NKT cells in vivo, suggesting that ILC2s are subject to a novel regulatory mechanism to mediate inflammation and tissue repair responses.
Materials and methods
Mice and reagents
C57BL/6 mice were purchased from CLEA Japan (Tokyo, Japan). Il5‐Venus knock‐in (Il5 +/Venus) mice18 and Il33 −/− mice39 on the C57BL/6 background were maintained under specific pathogen‐free conditions. Mice at 8–20 weeks of age were used for all experiments. Mice were handled in accordance with the Guidelines for Animal Experiments of the Research Institute, National Center for Global Health and Medicine. Interleukin‐33 and TSLP were purchased from R&D Systems (Minneapolis, MN); IL‐2, IL‐7 and IFN‐γ were purchased from PeproTech (Rocky Hill, NJ); and α‐galactosylceramide (α‐GalCer) was purchased from Funakoshi (Tokyo, Japan).
Lung and kidney cell preparation
Mice were injected intraperitoneally with recombinant mouse IL‐33 on days 0, 1 and 2 at a dose of 400 ng per mouse, and killed on day 3. Lungs were minced and digested in Hanks' balanced salt solution containing type I collagenase (1 mg/ml; Sigma‐Aldrich, St Louis, MO) and DNase I (200 U/ml; Roche Applied Science, Mannheim, Germany) for 1 hr at 37°. Tissue samples were next mashed through a 70‐µm cell strainer and washed with Hanks' balanced salt solution supplemented with 10% fetal calf serum. Red blood cells were removed by incubation for 2 min at room temperature in ammonium chloride (ACK) lysis buffer. Kidneys were minced and similarly digested in type I collagenase/DNase I for 1 hr at 37° with shaking, and then mashed through a 70‐µm cell strainer and washed with PBS. Kidney samples were suspended in 40% Percoll (GE Healthcare, Piscataway, NJ), under‐laid with 75% Percoll, and centrifuged at 800 g for 20 min at room temperature to isolate cells at interphase.
Flow cytometry and cell sorting
Single‐cell suspensions were pre‐incubated with 2.4G2 to block Fc receptors. Cells were stained with combinations of the following monoclonal antibodies: phycoerythrin‐conjugated anti‐CD25 (PC61; BioLegend, San Diego, CA), anti‐CD127 (SB/199; eBioscience, San Diego, CA) and anti‐ST2 (DIH9; BioLegend), allophycocyanin‐conjugated anti‐CD25 (PC61; BioLegend) and anti‐CD127 (A7R34; BioLegend), phycoerythrin‐Cy7‐conjugated anti‐Thy1.2 (30‐H12; BioLegend) and biotin‐conjugated anti‐CD3e (145‐2C11; Biolegend), anti‐CD4 (GK1.5; BioLegend), anti‐CD8 (53‐6.7; BioLegend), anti‐CD11b (M1/70; BioLegend), anti‐CD11c (N418; BioLegend), anti‐CD19 (6D5; BioLegend), anti‐B220 (RA3‐6B2; BioLegend), anti‐NK1.1 (PK136; BioLegend), anti‐T‐cell receptor‐γδ (GL3; BD Biosciences, San Jose, CA) and anti‐TER‐119 (TER‐119; Tonbo Biosciences, San Diego, CA) followed by allophycocyanin‐Cy7‐conjugated streptavidin. Dead cells were stained by addition of 7‐aminoactinomycin D (Sigma‐Aldrich). Identification of ILC2s was based on the expression of Thy1.2, CD25, CD127 and ST2 and the absence of lineage markers (Lin−). Sorting of ILC2 was performed with a FACSAria III (BD Biosciences), and phenotypic analysis was performed using a BD FACSCanto II (BD Biosciences). Data were analysed using flowjo (Tree Star Inc., San Carlos, CA).
Cell culture
Group 2 innate lymphoid cells were sorted from lung or kidney cell suspensions and cultured for 3 days in the presence or absence of IFN‐γ (10 ng/ml) in complete RPMI‐1640 medium supplemented with 10% fetal calf serum and 50 μm β‐mercaptoethanol at a density of 3 × 103 to 5 × 103 in 100 μl culture volume. Lung ILC2s were stimulated with IL‐33 (10 ng/ml) and TSLP (10 ng/ml; R&D Systems), and kidney ILC2s were stimulated with IL‐2 (10 ng/ml), IL‐7 (10 ng/ml), IL‐33 (10 ng/ml) and TSLP (10 ng/ml).
Quantitative RT‐PCR analysis
Total RNA was prepared from FACS‐sorted ILC2s using the RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations. The cDNA was synthesized using the High Capacity RNA‐to‐cDNA Kit (Applied Biosystems, Foster City, CA). Real‐time PCR was performed using TaqMan Fast Universal PCR Master Mix and primer/probe sets for the genes of interest in a StepOne Real‐Time PCR System (Applied Biosystems). The primers were as follows: Il5 (Mm00439646_m1), Il13 (Mm00434204_m1), Ifngr1 (Mm00599890_m1), Ifng (Mm01168134_m1) and Gapdh (Mm99999915_g1) (all Applied Biosystems). Target gene quantification was normalized to Gapdh expression.
SAGE‐seq analysis
Serial analysis of gene expression‐sequencing (SAGE‐seq) libraries were generated from total RNA of ILC2s using SOLiD SAGE Kit with Barcoding Adaptor Module and multiplexed using SOLiD RNA Barcoding Kit Module 1‐16 (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Briefly, total RNA was treated with DNase I, and then bound to Dynabeads Oligo(dT) EcoP magnetic beads for cDNA synthesis. The double‐stranded cDNA synthesized on the beads was digested with NlaIII and ligated to the adaptor containing an EcoP15I restriction enzyme recognition site. The adapter‐ligated cDNA was digested with EcoP15I to release the tag containing the adaptor sequence and 27 bp of unique sequence from a single transcript.
Sequencing
The size of the constructed library was quantified by the 2100 Bioanalyzer system with the High Sensitivity DNA kit (Agilent Technologies, Santa Clara, CA) and the SOLiD Library TaqMan Quantitation Kit (Life Technologies). Library DNA was subjected to emulsion PCR, enrichment, and deposition onto bead according to the supplier's protocol. Finally, DNA was sequenced using the SOLiD4 system (Life Technologies). All short reads from the SOLiD sequencer were aligned to mouse genome version 37 (mm9) using the bioscope program.
Cytokine measurements
Cytokine levels in supernatants were determined by Bio‐Plex Pro™ Mouse Cytokine 23‐plex Assay (Bio‐Rad Laboratories, Hercules, CA) in a Bio‐Plex 3D System (Bio‐Rad Laboratories) according to the manufacturer's instructions. Briefly, undiluted culture supernatant and standards were added to 96‐well microplates containing washed beads and incubated at room temperature for 1 hr with shaking. After washing, biotin‐labelled antibody was added to the wells. After 30 min of incubation at room temperature with shaking, streptavidin–phycoerythrin was added followed by 10 min of incubation with shaking. Data were collected with xPONENT for FLEXMAP 3D software, version 4.2 (Luminex Corporation, Austin, TX) and the data were analysed with bio‐plex manager 6.1 software (Luminex Corporation).
Interferon‐γ neutralization and α‐GalCer administration
Mice were given 500 µg of anti‐IFN‐γ monoclonal antibody (XMG1.2) intraperitoneally. Mice were given 100 µg/kg of α‐GalCer intraperitoneally 6 hr after treatment with anti‐IFN‐γ monoclonal antibody or PBS. The α‐GalCer was dissolved in PBS containing 5·6% sucrose, 0·75% l‐histidine and 0·5% Tween20 with heating at 80° for a few minutes, and then the solution was diluted in PBS. For in vivo treatment of Il33 −/− mice, 100 µg/kg of α‐GalCer or PBS was injected into mice intraperitoneally. On days 7, 8 and 9, 400 ng of IL‐33 was given to the α‐GalCer‐treated or PBS‐treated mice, and ILC2s were sort‐purified from lung and kidney suspensions on day 10.
Statistical analysis
Data were analysed using kaleidagraph (Synergy Software, Reading, PA). Unpaired two‐tailed Student's t‐test was used to determine significance. P values < 0·05 were considered statistically significant.
Results
ILC2s in lung express the receptor for IFN‐γ
To characterize IL‐5‐producing ILC2s in lung, we used Il5 +/Venus knock‐in mice in which Il5 expression is monitored by the expression of Venus, a modified green fluorescent protein.18 The ILC2s were identified as lineage negative (Lin; CD3ε, CD4, CD8, CD19, B220, CD11b, CD11c, NK1.1, T‐cell receptor‐γδ, TER119), Thy1.2+, CD25+ and CD127+ cells. Cell size and Venus expression were greatly increased in ILC2s from IL‐33‐injected Il5 +/Venus mice (34%, Fig. 1a, lower panels) compared with those from naive untreated mice (2·7%, Fig. 1a, upper panels). In addition to IL‐5, ILC2s produce IL‐13. Accordingly, the expression of Il5 and Il13 (Fig. 1b) was increased in ILC2s purified from the lungs of IL‐33‐injected mice.
Figure 1.
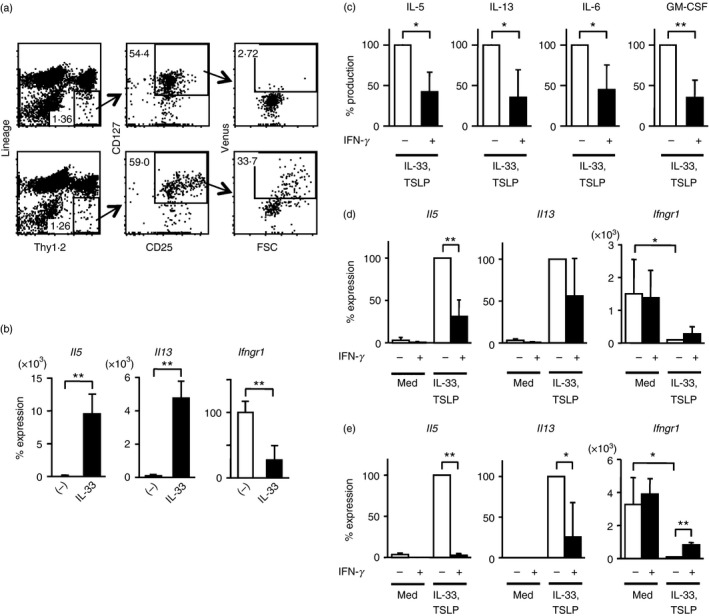
Interferon‐γ (IFN‐γ) suppresses production of interleukin‐5 (IL‐5), IL‐13, IL‐6 and granulocyte–macrophage colony‐stimulating factor (GM‐CSF) by activated group 2 innate lymphoid cells (ILC2s) in lung. (a) Flow cytometric analysis of leucocytes in lung obtained from naive Il5 +/Venus mice (top dot plots) or from Il5 +/Venus mice injected with recombinant IL‐33 (bottom dot plots). ILC2s were identified as Lin− Thy1.2+ CD25+ CD127+ cells, and IL‐5‐expressing activated ILC2s were large in cell size and positive for Venus expression. (b) Quantitative RT‐PCR analysis of Il5, Il13 and Ifngr1 mRNA in freshly isolated ILC2s from naive untreated mice (−) or mice injected with IL‐33 (IL‐33). The mRNA levels were normalized and presented as per cent maximum expression observed in ILC2s from naive untreated mice. (c, d) Purified ILC2s from IL‐33‐injected mice were cultured in the presence of IL‐33 and thymic stromal lymphopoietin (TSLP) with or without IFN‐γ for 3 days. Culture supernatants were collected and IL‐5, IL‐13, IL‐6 and GM‐CSF were quantified using a Bio‐Plex system (c). Cultured cells were harvested and Il5, Il13 and Ifngr1 mRNA expression was determined by quantitative RT‐PCR (d). (e) ILC2s purified from naive mice were cultured as in (d) for 3 days and the expression of Il5, Il13 and Ifngr1 mRNA was determined by quantitative RT‐PCR. The mRNA levels were normalized by Gapdh mRNA expression and indicated as percent maximum expression found in ILC2s cultured with IL‐33 and TSLP (d, e). Data shown represent mean ± SD of three or four independent experiments, *P < 0·05, **P < 0·01 (two‐tailed Student's t‐test) (a–e).
To examine the regulation operating in ILC2s, we studied the gene expression profile of IL‐5‐producing Venus+ ILC2s sort‐purified from the lungs of IL‐33‐treated mice by SAGE. We found that Arg1 was one of the frequently detected genes (Table 1), consistent with a previous report showing that ILC2s express high amounts of arginase‐1.40 We noticed that Ifngr1, the receptor for the signature cytokine of Th1 cells, was also among the top 25 protein‐coding genes frequently detected in our SAGE of IL‐5‐producing ILC2s (Table 1). We confirmed transcript expression by RT‐PCR, and found that the levels of Ifngr1 were higher in ILC2s obtained from naive mice than in activated ILC2s from IL‐33‐injected mice (Fig. 1b).
Table 1.
Top 25 protein‐coding genes detected by serial analysis of gene expression of lung interleukin‐5 (IL‐5) ‐producing innate lymphoid cells obtained from IL‐33 injected mice
| Gene | Description | Detection count |
|---|---|---|
| Arg1 | Arginase, liver | 12 480 |
| Hnrnpl | Heterogeneous nuclear ribonucleoprotein | 4219 |
| Hnrnpa1 | Heterogeneous nuclear ribonucleoprotein A1 | 4028 |
| Hsp90aa1 | Heat‐shock protein 90, α (cytosolic), class A member 1 | 3352 |
| Rpl4 | Ribosomal protein L4 | 3292 |
| Ifngr1 | Interferon‐γ receptor 1 | 2733 |
| Glul | Glutamate‐ammonia ligase (glutamine synthetase) | 2620 |
| Dapk1 | Death‐associated protein kinase 1 | 2510 |
| Lmnb1 | Lamin B1 | 2372 |
| Txnrd1 | Thioredoxin reductase 1 | 2167 |
| Capza1 | Capping protein (actin filament) muscle Z‐line, α1 | 2145 |
| Atp5 g3 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 | 2105 |
| Ppp1ca | Protein phosphatase 1, catalytic subunit, α isoform | 1997 |
| Prr13 | Proline rich 13 | 1972 |
| H3f3b | H3 histone, family 3B | 1868 |
| Rpl38 | Ribosomal protein L38 | 1847 |
| Hspa8 | Heat‐shock protein 8 | 1640 |
| Fgl2 | Fibrinogen‐like protein 2 | 1599 |
| Dazap2 | DAZ‐associated protein 2 | 1597 |
| Cd81 | CD81 antigen | 1569 |
| Atp5a1 | ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit 1 | 1562 |
| Gm9843 | Predicted gene 9843 | 1530 |
| Cxcl2 | Chemokine (C‐X‐C motif) ligand 2 (MIP2) | 1502 |
| Hnrnpa2b1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | 1476 |
| Atp5b | ATP synthase, H+ transporting mitochondrial F1 complex, β subunit | 1436 |
Interferon‐γ suppresses cytokine production by ILC2s
Interferon‐γ produced by Th1 cells suppresses the expansion and development of Th2 cells from naive Th0 cells.29 We wanted to determine if IFN‐γ affects the function of ILC2s, which normally produce high levels of Th2 cytokines, similar to the suppressive effect on Th2 cells. We purified ILC2s from IL‐33‐injected mice and cultivated them with IL‐33 and TSLP in the presence or absence of IFN‐γ. Production of IL‐5 and IL‐13 at the protein (Fig. 1c) and mRNA (Fig. 1d) levels was significantly inhibited in the presence of IFN‐γ. Interleukin‐6 and granulocyte–macrophage colony‐stimulating factor produced by ILC2s were also suppressed by IFN‐γ (Fig. 1c). Consistent with the higher expression of Ifngr1 transcripts in ILC2s from naive mice compared with those from IL‐33‐injected mice (Fig. 1b), IFN‐γ significantly inhibited the expression of IL‐5 and IL‐13 in ILC2s from naive mice after stimulation with IL‐33 and TSLP (Fig. 1e). Cultivating ILC2s from IL‐33‐injected mice or naive mice in the presence of IL‐33 resulted in the down‐regulation of IFN‐γ receptor (Fig. 1d, e). In naive ILC2s, IFN‐γ partially but significantly prevented the down‐regulation of IFN‐γ receptor induced by IL‐33 and TSLP (Fig. 1e). Hence, IFN‐γ suppressed cytokine production by ILC2s in lung, and ILC2s in naive mice were more susceptible to the IFN‐γ‐mediated suppression compared with ILC2s activated by IL‐33, which resulted in the down‐regulation of Ifngr1.
Interferon‐γ decreased cytokine production by kidney ILC2s
We next examined how kidney‐resident ILC2s respond to IFN‐γ. We identified Lin− Thy1.2+ CD25+ CD127+ ILC2s, only a few of which were Venus+ (~10%), IL‐5‐producing ILC2s in the kidneys of naive mice (Fig. 2a). To support growth or survival of small numbers of purified kidney ILC2s in culture, we added IL‐2 and IL‐7, known growth factors for ILC2s in fat or lung, in combination with IL‐33 and TSLP. Cultivation of purified ILC2s from kidneys with IL‐2, IL‐7, IL‐33 and TSLP resulted in robust IL‐5 production indicated by the increased fraction (~90%) of Venus+ cells and the high intensity of Venus expression (Fig. 2b, left plot). The addition of IFN‐γ into the culture decreased the frequency of Venus+ ILC2s (~65%), as well as reducing the intensity of Venus expression (Fig. 2b, right plot). Expression of ST2, a component of the IL‐33 receptor, was also significantly down‐regulated following stimulation with IFN‐γ (Fig. 2b). Il5 and Il13 transcripts induced by IL‐2, IL‐7, IL‐33 and TSLP stimulation were also measured by RT‐PCR, and the presence of IFN‐γ significantly suppressed the expression of Il5 and Il13 in ILC2s (Fig. 2c). Hence, IFN‐γ inhibits cytokine production by kidney ILC2s as well as lung ILC2s.
Figure 2.
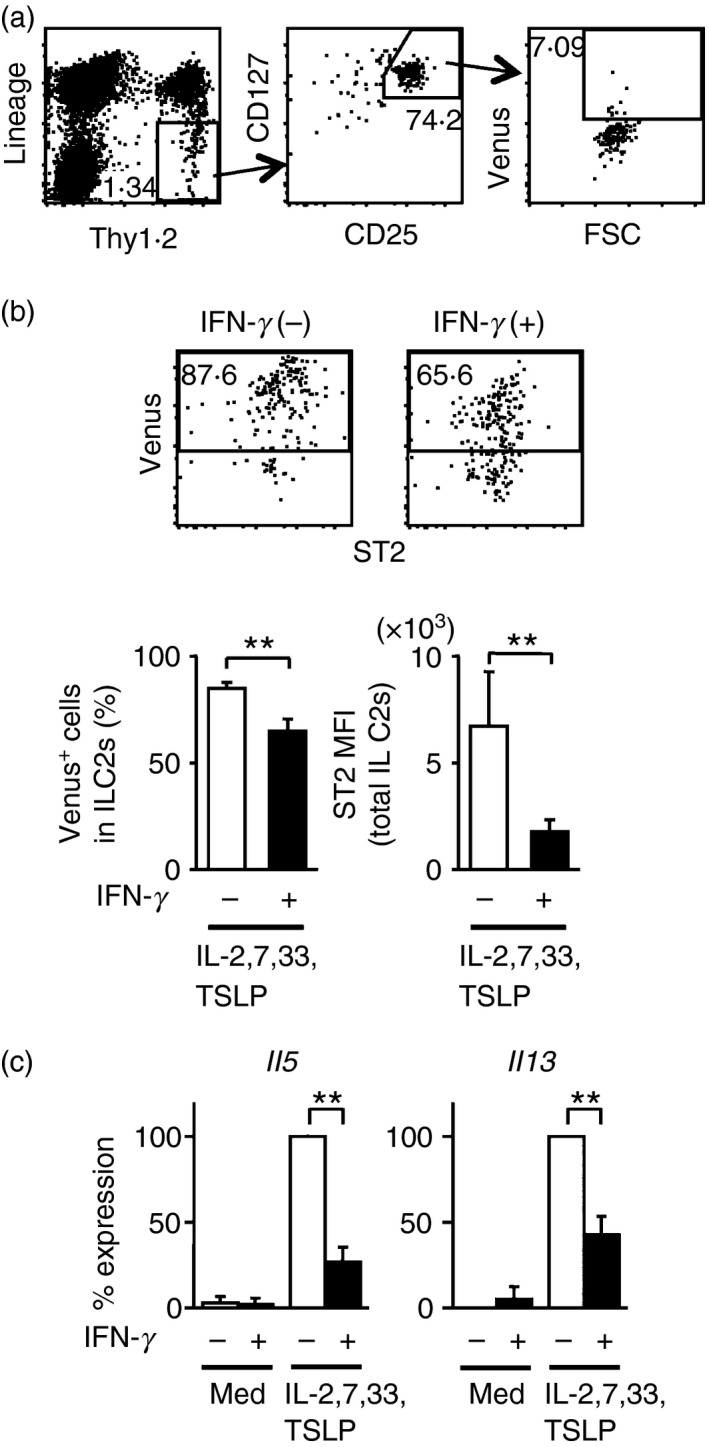
Interferon‐γ (IFN‐γ) inhibits interleukin‐5 (IL‐5) and IL‐13 production by kidney‐resident group 2 innate lymphoid cells (ILC2s). (a) Flow cytometric analysis of ILC2s from the kidneys of naive Il5 +/Venus mice. (b) Sorted ILC2s were cultured with IL‐2, IL‐7, IL‐33 and thymic stromal lymphopoietin (TSLP) in the presence or absence of IFN‐γ, and Venus and ST2 expression was analysed by flow cytometry. Shown are representative plots (upper panels) and the percentage of Venus+ cells and mean fluorescence intensity (MFI) of ST2 expression (mean ± SD) in the ILC2 fraction (lower graphs) obtained from independent four experiments. (c) Quantitative RT‐PCR analysis for Il5 and Il13 mRNA in cultured ILC2s from the kidneys of naive mice. Results are presented as per cent maximum expression observed in ILC2s stimulated by IL‐2, IL‐7, IL‐33 and TSLP. Data shown are the mean ± SD of four independent experiments, **P < 0·01.
Interferon‐γ constrains ILC2 cytokine production induced after NKT cell activation
To examine the negative regulation of ILC2s by IFN‐γ in vivo, we employed a model of ILC2 activation mediated by NKT cells. NKT cells produce IL‐33 upon viral infection and are involved in the activation of ILC2s.23 The glycolipid ligand α‐galactosylceramide (α‐GalCer) specifically activates NKT cells41 to produce IFN‐γ and IL‐33.23 First, we investigated whether IL‐5 and IL‐13 production by ILC2s after NKT cell activation relied upon IFN‐γ.
Mice were injected with α‐GalCer with or without anti‐IFN‐γ. Two days later, ILC2s in lungs or kidneys were isolated and the expression of IL‐5 and IL‐13 was examined. We observed elevated expression of IFN‐γ transcripts and increased numbers of NKT cells in the lungs of α‐GalCer‐treated mice (data not shown). Neutralization of IFN‐γ resulted in augmented expression of IL‐5 (2·0‐fold) and IL‐13 (1·9‐fold) in lung ILC2s (Fig. 3a). The impact of IFN‐γ blockade was more remarkable in kidney ILC2s for IL‐5 (4·2‐fold) and IL‐13 (2·3‐fold) expression (Fig. 3b). These results demonstrate that IFN‐γ, together with alarmins in certain circumstances, inhibits cytokine production by ILC2s in vivo.
Figure 3.
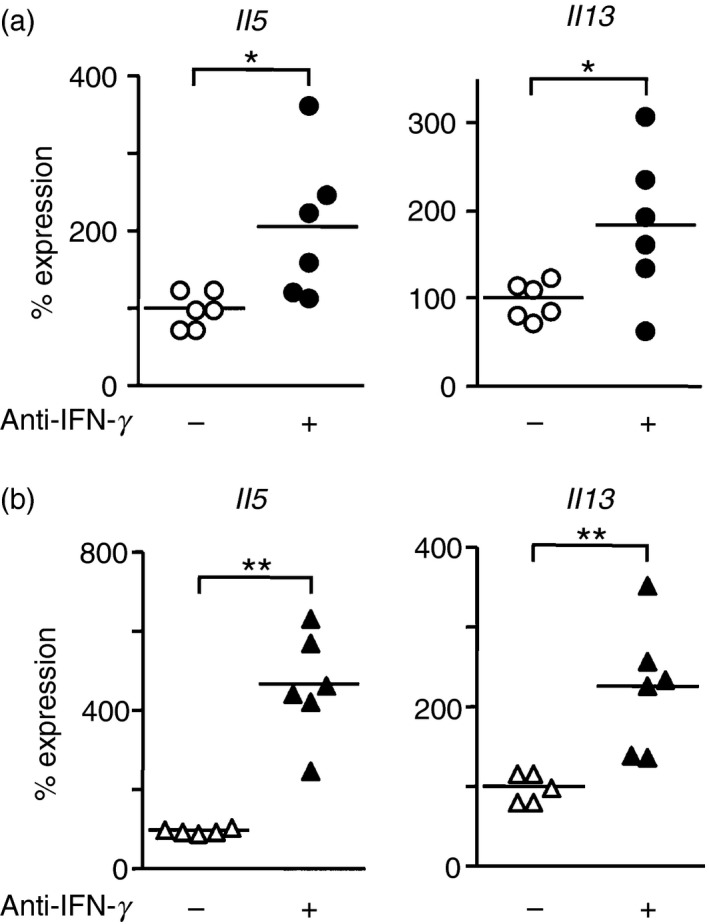
Neutralization of interferon‐γ (IFN‐γ) by anti‐IFN‐γ antibodies results in augmented expression of interleukin‐5 (IL‐5) and IL‐13 in group 2 innate lymphoid cells (ILC2s) obtained from lungs and kidneys of mice injected with α‐galactosylceramide (α‐GalCer). Mice were intraperitoneally (i.p.) injected with 500 µg of anti‐IFN‐γ (+) or PBS (−). Six hours later, mice were further i.p. injected with 100 μg/kg of α‐GalCer. Two days after the α‐GalCer injection, ILC2s were sort‐purified from lungs (a) or kidneys (b) and the mRNA levels for IL‐5 and IL‐13 were determined by quantitative RT‐PCR. Data shown are combined results obtained from five or six independent experiments. Each symbol represents individual mice, and bars represent the mean of each group of mice. *P < 0·05, **P < 0·01.
Exposure to IFN‐γ before activation reduces ILC2 cytokine production
Next, we investigated the effect of IFN‐γ present in the microenvironment before the activation of ILC2s. We injected α‐GalCer into Il33 −/− mice in which NKT cells produced IFN‐γ but not IL‐33. Seven days later, ILC2s were activated by administration of exogenous IL‐33, and ILC2 cytokine production was measured. Injection of recombinant IL‐33 resulted in increased expression of Il5 and Il13 in ILC2s harvested from the lungs and kidneys of Il33 −/− mice (Fig. 4a and b). In contrast, Il33 −/− mice pre‐treated with α‐GalCer showed increased Ifng expression in lung (Fig. 4c), and marked suppression of Il5 and Il13 transcripts in both lung and kidney ILC2s after injection of exogenous IL‐33 (Fig. 4a, b). These observations demonstrate that IFN‐γ pre‐existing in tissues suppressed ILC2 cytokine production after activation by IL‐33.
Figure 4.
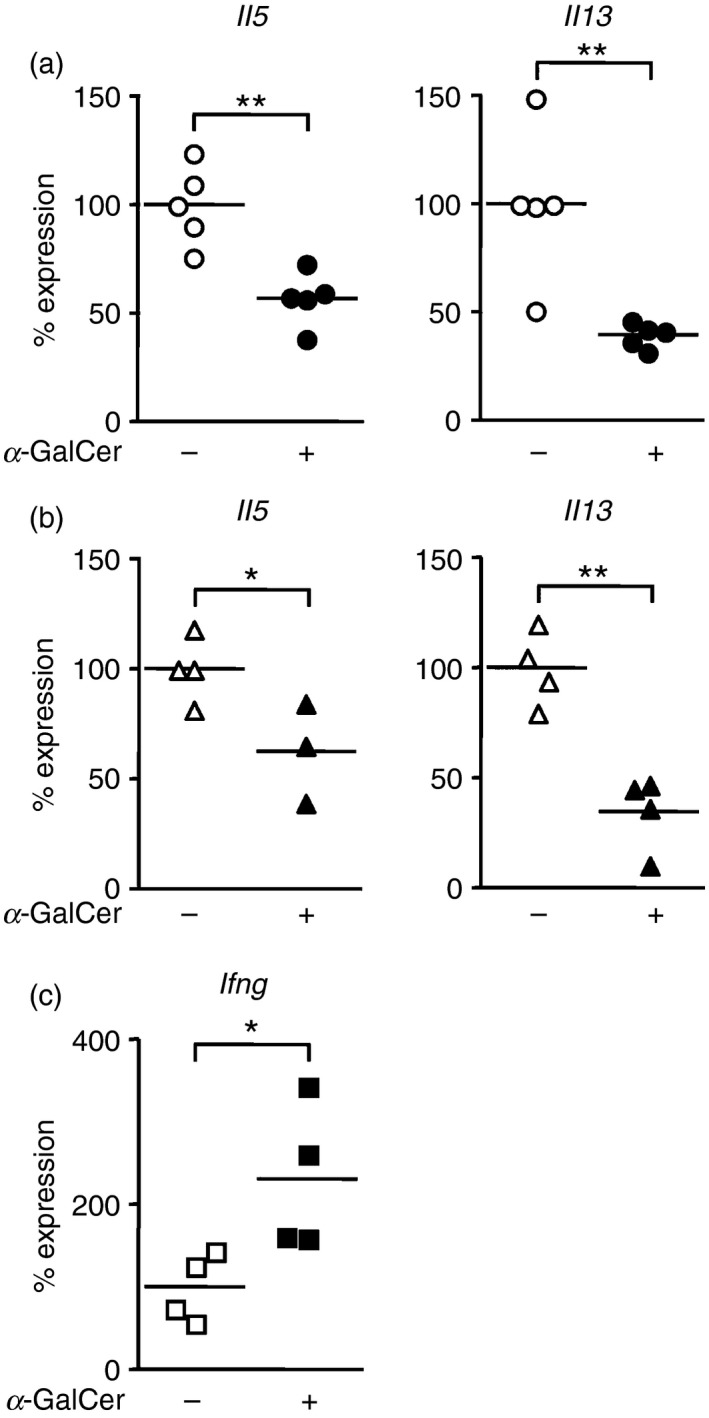
Pre‐existing interferon‐γ (IFN‐γ) reduces cytokine production from group 2 innate lymphoid cells (ILC2s). Il33 −/− mice were injected intraperitoneally (i.p.) with 100 μg/kg of α‐galactosylceramide (α‐GalCer) (+) or PBS (−) on day 0. Seven days later, recombinant interleukin‐33 (IL‐33) was administered to the Il33 −/− mice at a dose of 400 ng per injection per day, for three consecutive days on days 7, 8 and 9. ILC2s were sort‐purified from lungs (a) and kidneys (b) and the mRNA levels of IL‐5 and IL‐13 were determined by quantitative RT‐PCR on day 10. Ifng expression in total lung cells was also measured by quantitative RT‐PCR (c). Data shown are combined results obtained from of three to five independent experiments. Each symbol represents individual mice, and bars represent the mean of each group of mice. *P < 0·05, **P < 0·01.
Discussion
Type 2 immune responses are characterized by IL‐4, IL‐5, IL‐9 and IL‐13, and are important for protecting the host against helminth infection, suppression of type 1‐driven autoimmune disease, neutralizing toxins, and regulating tissue repair.2, 42 ILC2s produce a large amount of Th2 cytokines and promote type 2 immune responses. In this study, we found that IFN‐γ receptor (Ifngr1) was expressed on IL‐33‐activated lung ILC2s. Interferon‐γ is a Th1 cytokine that induces type 1 immune responses and suppresses type 2 immunity by suppressing the development and expansion of Th2 cells.30 Our results demonstrated that IFN‐γ also inhibited ILC2s in addition to Th2 cells, and directly suppressed the production of Th2 cytokines such as IL‐5, IL‐13, IL‐6 and granulocyte–macrophage colony‐stimulating factor from ILC2s.
The suppressive effects of IFN‐γ on ILC2s were confirmed in two in vivo models. First, we analysed IL‐5 and IL‐13 production by ILC2s using α‐GalCer in the presence of blocking antibodies specific for IFN‐γ. α‐GalCer is a potent and specific activator of NKT cells41, 43 and induces IFN‐γ and IL‐4 production by NKT cells.36 NKT cells are also a source of IL‐33 that activates ILC2s during viral infection.23 Injection of α‐GalCer into mice resulted in the activation of NKT cells and production of NKT cell‐derived IL‐33, which led to the activation of ILC2s. The blockade of IFN‐γ, the primary source of which was NKT cells activated by α‐GalCer, significantly enhanced IL‐5 and IL‐13 production by ILC2s, indicating that IFN‐γ inhibits ILC2 function. Second, we use Il33 −/− mice to examine the effect of pre‐existing IFN‐γ on ILC2 activation. In contrast to IFN‐γ blockade, pre‐existing IFN‐γ, derived from α‐GalCer‐stimulated Il33‐deficient NKT cells, markedly suppressed IL‐5 and IL‐13 production by ILC2s activated with recombinant IL‐33. The inhibitory effects of IFN‐γ on ILC2s were observed in both lung and kidney ILC2s.
During this study, Molofsky and others showed that IL‐33 signalling is required for the development and maintenance of regulatory T cells, and that IFN‐γ regulates ILC2 activation and limits regulatory T‐cell proliferation in visceral adipose tissue.44, 45 They also showed that co‐infection of Listeria monocytogenes, which elicits IFN‐γ production by CD8+ cells, and helminths that induce IL‐33, resulted in the inhibition of ILC2 activation and regulatory T‐cell expansion. We have independently demonstrated the effects of NKT cell‐derived IFN‐γ induced in vivo by α‐GalCer administration and pre‐existing in tissues before activation of ILC2s by IL‐33.
Expression of Ifngr1 on ILC2s was down‐regulated by in vivo stimulation with IL‐33. Accordingly, the inhibitory effects of IFN‐γ were greater in naive ILC2s. Interferon‐γ receptor down‐regulation was mainly the result of the direct effect of IL‐33 on ILC2s because it was also observed during the activation of ILC2s by IL‐33 in cultures of lung ILC2s (Fig. 1d, e). Interferon‐γ is primarily produced by Th1 cells, CD8+ effector T cells, NK cells and activated NKT cells.46 The activation of ILC2s is inhibited in Th1‐skewed microenvironments, as are Th2 immune responses. As schematically presented in Figure 5, IFN‐γ inhibits the differentiation of Th2 cells and also prohibits the production of IL‐5 and IL‐13 by ILC2s activated by epithelial‐derived IL‐33 and TSLP following exposure to allergens, or NKT cell‐derived IL‐33 upon viral infection or exposure to NKT‐activating ligands such as α‐GalCer. It appears that the suppression of ILC2s by IFN‐γ occurs during the reciprocal regulation between Th1 and Th2 immune responses. Exposure to IL‐33 down‐regulated Ifngr1 expression, which resulted in reduced inhibition of ILC2 function by IFN‐γ. In other words, IL‐33 activates ILC2s and makes them somewhat resistant to IFN‐γ‐mediated inhibition. Preceding exposure of ILC2s to IL‐33 or IFN‐γ severely impacts downstream ILC2 functions.
Figure 5.
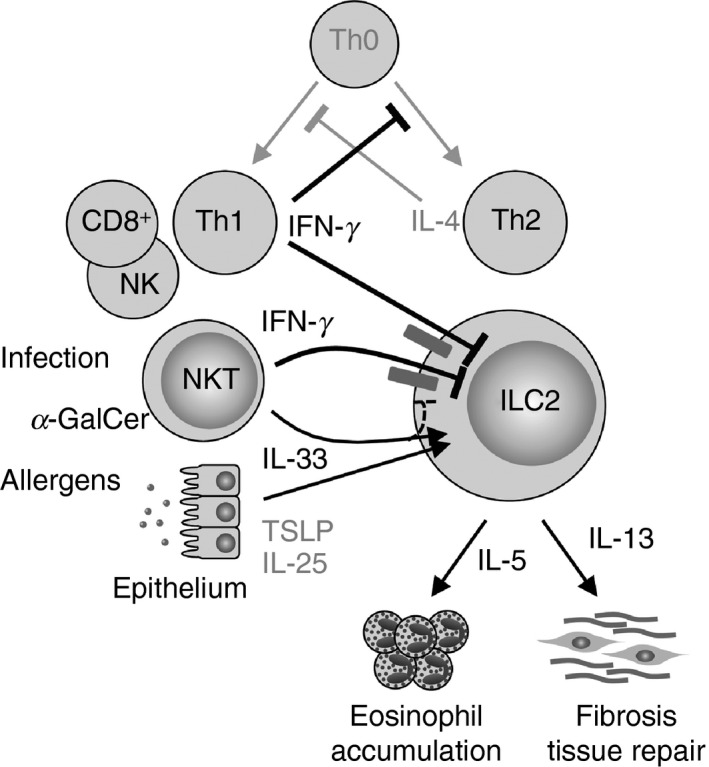
Schematic model for the regulation of group 2 innate lymphoid cell (ILC2) cytokine production by interferon‐γ (IFN‐γ). IFN‐γ derived from T helper type 1 (Th1), natural killer T (NKT), NK or CD8+ effector T cells inhibits the differentiation and function of Th2 cells. IFN‐γ also limits interleukin‐5 (IL‐5) and IL‐13 production by ILC2s that are activated by epithelial‐derived IL‐33 and thymic stromal lymphopoietin (TSLP) following exposure to allergens, by NKT‐cell‐derived IL‐33 upon viral infection, or exposure to NKT‐activating ligands such as α‐galactosylceramide (α‐GalCer).
In conclusion, our findings demonstrate that IFN‐γ directly inhibits IL‐5 and IL‐13 production by ILC2s, and that blocking IFN‐γ restored the ability of activated ILC2s to produce IL‐5 and IL‐13. Interferon‐γ antagonizes ILC2 function in a way that is similar to that in which IFN‐γ impacts Th2 cells and constrains type 2 immune responses.
Disclosure
All authors declare that no competing financial interests exist.
Acknowledgements
We thank our colleagues for helpful discussions and for technical assistance. This work was supported by JSPS KAKENHI Grant Number 25293097 (S.T.), and by The Grants for National Center for Global Health and Medicine (25‐107, 25‐103, 25‐104, 26‐110) (S.T.).
References
- 1. von Moltke J, Locksley RM. I‐L‐C‐2 it: type 2 immunity and group 2 innate lymphoid cells in homeostasis. Curr Opin Immunol 2014; 31:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wynn TA. Type 2 cytokines: mechanisms and therapeutic strategies. Nat Rev Immunol 2015; 15:271–82. [DOI] [PubMed] [Google Scholar]
- 3. Licona‐Limon P, Kim LK, Palm NW, Flavell RA. TH2, allergy and group 2 innate lymphoid cells. Nat Immunol 2013; 14:536–42. [DOI] [PubMed] [Google Scholar]
- 4. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G et al Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol 2013; 13:145–9. [DOI] [PubMed] [Google Scholar]
- 5. Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells – how did we miss them? Nat Rev Immunol 2013; 13:75–87. [DOI] [PubMed] [Google Scholar]
- 6. McHedlidze T, Waldner M, Zopf S, Walker J, Rankin AL, Schuchmann M et al Interleukin‐33‐dependent innate lymphoid cells mediate hepatic fibrosis. Immunity 2013; 39:357–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Molofsky AB, Nussbaum JC, Liang HE, Van Dyken SJ, Cheng LE, Mohapatra A et al Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J Exp Med 2013; 210:535–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H et al Innate production of TH2 cytokines by adipose tissue‐associated c‐Kit+ Sca‐1+ lymphoid cells. Nature 2010; 463:540–4. [DOI] [PubMed] [Google Scholar]
- 9. Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB et al Type 2 innate lymphoid cells control eosinophil homeostasis. Nature 2013; 502:245–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA et al Innate lymphoid cells promote lung‐tissue homeostasis after infection with influenza virus. Nat Immunol 2011; 12:1045–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mjosberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B et al Human IL‐25‐ and IL‐33‐responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol 2011; 12:1055–62. [DOI] [PubMed] [Google Scholar]
- 12. Halim TY, Steer CA, Matha L, Gold MJ, Martinez‐Gonzalez I, McNagny KM et al Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell‐mediated allergic lung inflammation. Immunity 2014; 40:425–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van Dyken SJ, Mohapatra A, Nussbaum JC, Molofsky AB, Thornton EE, Ziegler SF et al Chitin activates parallel immune modules that direct distinct inflammatory responses via innate lymphoid type 2 and γδ T cells. Immunity 2014; 40:414–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med 2012; 18:716–25. [DOI] [PubMed] [Google Scholar]
- 15. Takatsu K, Nakajima H. IL‐5 and eosinophilia. Curr Opin Immunol 2008; 20:288–94. [DOI] [PubMed] [Google Scholar]
- 16. Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol 2013; 13:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Allen JE, Sutherland TE. Host protective roles of type 2 immunity: parasite killing and tissue repair, flip sides of the same coin. Semin Immunol 2014; 26:329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ikutani M, Yanagibashi T, Ogasawara M, Tsuneyama K, Yamamoto S, Hattori Y et al Identification of innate IL‐5‐producing cells and their role in lung eosinophil regulation and antitumor immunity. J Immunol 2012; 188:703–13. [DOI] [PubMed] [Google Scholar]
- 19. Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science 2015; 348:aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zaiss DM, Gause WC, Osborne LC, Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity 2015; 42:216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell‐type cytokines in protease allergen‐induced airway inflammation. Immunity 2012; 36:451–63. [DOI] [PubMed] [Google Scholar]
- 22. Chang YJ, Kim HY, Albacker LA, Baumgarth N, McKenzie AN, Smith DE et al Innate lymphoid cells mediate influenza‐induced airway hyper‐reactivity independently of adaptive immunity. Nat Immunol 2011; 12:631–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gorski SA, Hahn YS, Braciale TJ. Group 2 innate lymphoid cell production of IL‐5 is regulated by NKT cells during influenza virus infection. PLoS Pathog 2013; 9:e1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilhelm C, Hirota K, Stieglitz B, Van Snick J, Tolaini M, Lahl K et al An IL‐9 fate reporter demonstrates the induction of an innate IL‐9 response in lung inflammation. Nat Immunol 2011; 12:1071–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim BS, Wang K, Siracusa MC, Saenz SA, Brestoff JR, Monticelli LA et al Basophils promote innate lymphoid cell responses in inflamed skin. J Immunol 2014; 193:3717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xue L, Salimi M, Panse I, Mjosberg JM, McKenzie AN, Spits H et al Prostaglandin D2 activates group 2 innate lymphoid cells through chemoattractant receptor‐homologous molecule expressed on TH2 cells. J Allergy Clin Immunol 2014; 133:1184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Doherty TA, Khorram N, Lund S, Mehta AK, Croft M, Broide DH. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J Allergy Clin Immunol 2013; 132:205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maazi H, Patel N, Sankaranarayanan I, Suzuki Y, Rigas D, Soroosh P et al ICOS : ICOS‐ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity 2015; 42:538–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mosmann TR, Sad S. The expanding universe of T‐cell subsets: Th1, Th2 and more. Immunol Today 1996; 17:138–46. [DOI] [PubMed] [Google Scholar]
- 30. Erb KJ, Holloway JW, Sobeck A, Moll H, Le Gros G. Infection of mice with Mycobacterium bovis–Bacillus Calmette–Guérin (BCG) suppresses allergen‐induced airway eosinophilia. J Exp Med 1998; 187:561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M et al T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis . Immunity 2007; 27:505–17. [DOI] [PubMed] [Google Scholar]
- 32. Szabo SJ, Jacobson NG, Dighe AS, Gubler U, Murphy KM. Developmental commitment to the Th2 lineage by extinction of IL‐12 signaling. Immunity 1995; 2:665–75. [DOI] [PubMed] [Google Scholar]
- 33. Martin‐Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A et al Induced recruitment of NK cells to lymph nodes provides IFN‐γ for TH1 priming. Nat Immunol 2004; 5:1260–5. [DOI] [PubMed] [Google Scholar]
- 34. Sad S, Marcotte R, Mosmann TR. Cytokine‐induced differentiation of precursor mouse CD8+ T cells into cytotoxic CD8+ T cells secreting Th1 or Th2 cytokines. Immunity 1995; 2:271–9. [DOI] [PubMed] [Google Scholar]
- 35. Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon‐γ: an overview of signals, mechanisms and functions. J Leukoc Biol 2004; 75:163–89. [DOI] [PubMed] [Google Scholar]
- 36. Taniguchi M, Seino K, Nakayama T. The NKT cell system: bridging innate and acquired immunity. Nat Immunol 2003; 4:1164–5. [DOI] [PubMed] [Google Scholar]
- 37. Else KJ, Finkelman FD, Maliszewski CR, Grencis RK. Cytokine‐mediated regulation of chronic intestinal helminth infection. J Exp Med 1994; 179:347–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oldroyd SD, Thomas GL, Gabbiani G, El Nahas AM. Interferon‐γ inhibits experimental renal fibrosis. Kidney Int 1999; 56:2116–27. [DOI] [PubMed] [Google Scholar]
- 39. Oboki K, Ohno T, Kajiwara N, Arae K, Morita H, Ishii A et al IL‐33 is a crucial amplifier of innate rather than acquired immunity. Proc Natl Acad Sci U S A 2010; 107:18581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bando JK, Nussbaum JC, Liang HE, Locksley RM. Type 2 innate lymphoid cells constitutively express arginase‐I in the naive and inflamed lung. J Leukoc Biol 2013; 94:877–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Godfrey DI, Rossjohn J. New ways to turn on NKT cells. J Exp Med 2011; 208:1121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gause WC, Wynn TA, Allen JE. Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat Rev Immunol 2013; 13:607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Venkataswamy MM, Porcelli SA. Lipid and glycolipid antigens of CD1d‐restricted natural killer T cells. Semin Immunol 2010; 22:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Molofsky AB, Van Gool F, Liang HE, Van Dyken SJ, Nussbaum JC, Lee J et al Interleukin‐33 and interferon‐γ counter‐regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity 2015; 43:161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vasanthakumar A, Moro K, Xin A, Liao Y, Gloury R, Kawamoto S et al The transcriptional regulators IRF4, BATF and IL‐33 orchestrate development and maintenance of adipose tissue‐resident regulatory T cells. Nat Immunol 2015; 16:276–85. [DOI] [PubMed] [Google Scholar]
- 46. Bourgeois E, Van LP, Samson M, Diem S, Barra A, Roga S et al The pro‐Th2 cytokine IL‐33 directly interacts with invariant NKT and NK cells to induce IFN‐γ production. Eur J Immunol 2009; 39:1046–55. [DOI] [PubMed] [Google Scholar]


