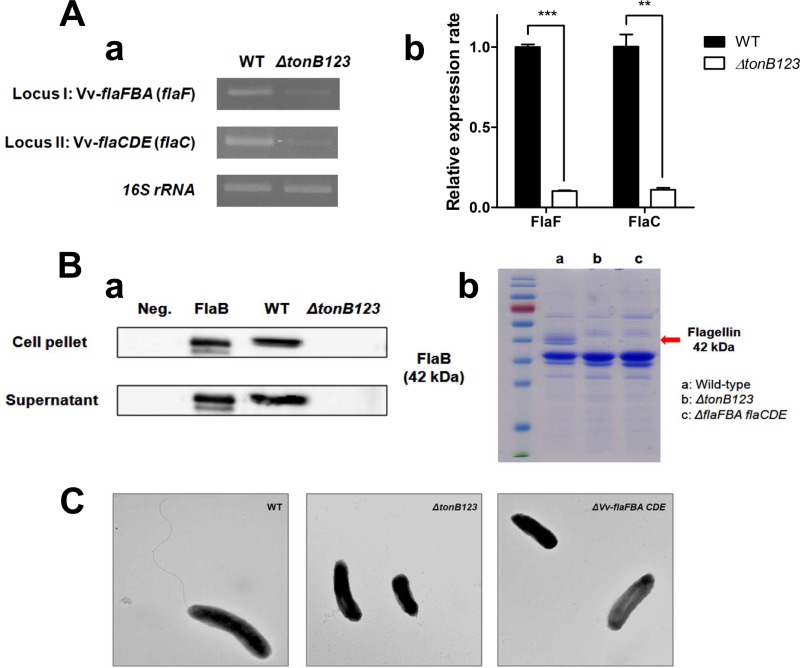
FIG 7.
Defects of the transcription (A) and translation (B) of flagellin genes and loss of flagellation in the tonB123 triple mutant (C). (A) RNA was isolated from log-phase bacterial cells that had been cultured in the rat peritoneal cavity and was then converted to cDNA. Conventional (a) and real-time (b) RT-PCR was performed using primers specific for flaF and flaC, as shown in Table S2 in the supplemental material. The error bars represent the standard errors of means from three independent experiments. Statistical analysis was carried out using Student's t test (**, P < 0.01, and ***, P < 0.001, compared to the wild-type strain). (B) (a) Bacteria were cultured in 2.5% NaCl HI to log-phase growth, and then Western blot analysis was subsequently carried out in the V. vulnificus cell pellet and culture supernatant using an anti-FlaB antibody (a band of approximately 42 kDa). The ΔflaFBA flaCDE strain was used as a negative control (Neg.). (b) The OMP preparations were analyzed by 12% SDS-PAGE. Two major OMP bands were missing in the ΔtonB123 mutant compared to the wild-type strain, which were subsequently identified as flagellin proteins (42 kDa) using MALDI-TOF MS. (C) Electron micrograph of the log-phase V. vulnificus strains under ×25,000 magnification.