Figure 1.
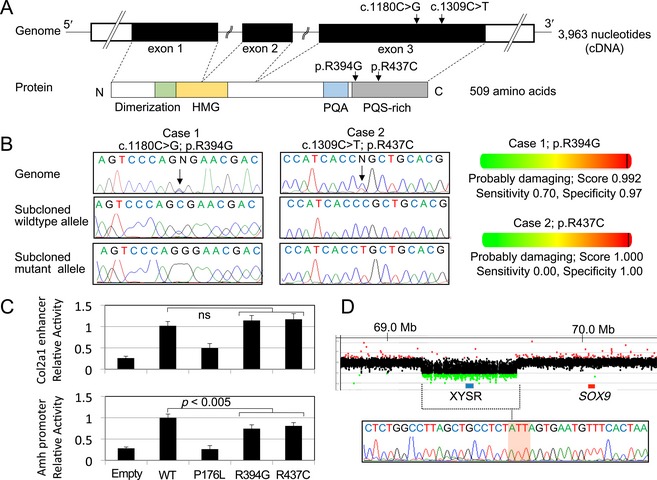
SOX9 abnormalities in patients 1–3. (A) Genomic and protein structures of SOX9/SOX9. The positions of the c.1180C>G (p.Arg394Gly) and c.1309C>T (p.Arg437Cys) mutations are indicated by arrows. White and black boxes in the upper panel indicate the untranslated and coding regions, respectively. Colored boxes in the lower panel indicate dimerization (codon 60–101) (Bernard et al. 2003), high‐mobility group (HMG: codon 101–184), proline/glutamine/alanine (PQA: codon 339–379), and proline/glutamine/serine‐rich (PQS‐rich: codon 386–509) domains (McDowall et al. 1999). (B) Nucleotide substitutions detected in patients 1 and 2. Left panel: electro chromatograms of the mutations. The mutated nucleotides are indicated by arrows. Right panel: in silico functional prediction of mutant proteins. (C) In vitro assays using reporters containing the Col2a1 enhancer or Amh promoter. The transactivating activity of p.Arg394Gly and p.Arg437Cys mutants was compared to that of the known ACD‐associated SOX9 mutant p.Pro176Leu (Michel‐Calemard et al. 2004). The results are expressed as the mean ± one standard deviation. Relative transactivating activities of the SOX9 mutants against the wild‐type are shown. Empty: empty expression vector; ns: not significant. (D) SOX9 upstream deletion in patient 3. Upper panel: array‐based comparative genomic hybridization analysis. The black, red, and green dots denote signals indicative of the normal, increased (> +0.5) and decreased (< −1.0) copy‐numbers, respectively. The blue and red boxes represent previously reported XY sex reversal region (XYSR) (Kim et al. 2015) and SOX9 exons, respectively. Genomic positions refer to the UCSC database (http://genome.ucsc.edu/; GRCh37/hg19). Lower panel: sequence of the fusion junction. The junction is accompanied by a short‐nucleotide insertion of unknown origin (the red‐shaded area).
