Figure 1.
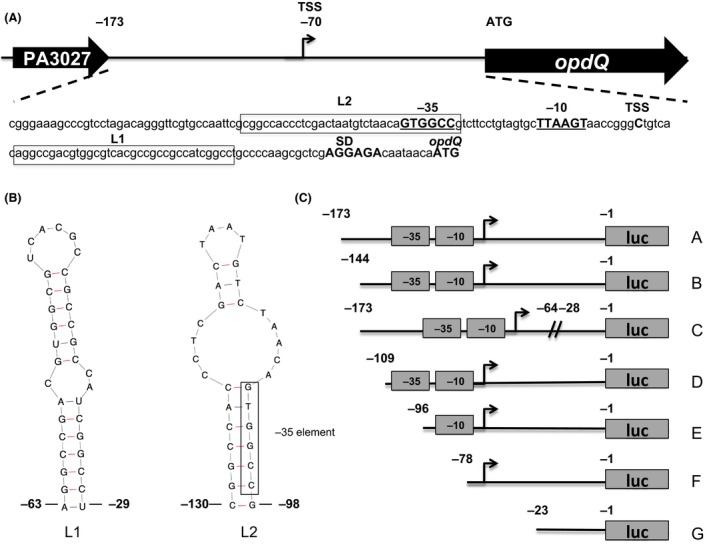
Mapping the opdQ promoter sequences in Pseudomonas aeruginosa. (A) Genetic organization of the opdQ structural gene and its promoter region. 5′ rapid amplification of cDNA ends and sequence analysis revealed transcription of opdQ in P. aeruginosa initiates from a single start site (TSS) at a cytosine residue 70 nucleotides upstream of the ATG of the opdQ structural gene. The putative ‐10 and ‐35 elements are indicated (underlined and bolded), whereas SD indicates the putative Shine–Dalgarno sequence. Boxed sequences designated L1 and L2 are nucleotides that form putative stem loops. (B) Predicted nucleic acid secondary structure of the PA3037‐opdQ intergenic region containing the opdQ promoter region showing nucleotide positions that form the L1 and L2 stem‐loop structures. These predicted structures represent the boxed sequences in (A). (C) Promoter::luciferase fusion clones representing different lengths of the opdQ promoter and its upstream sequences were fused to a luciferase reporter gene (luc) from vector pSPluc+ and subcloned into the broad‐host range vector pMP220. The opdQ promoter sequences (DNA fragments) and the putative ‐10 and ‐35 elements are indicated. These clones included sequences of the entire PA3037‐opdQ intergenic region (clone A), the removal of sequences upstream of the ‐35 element (clone B), an internal deletion of the L1 loop within the opdQ UTR (clone C), the removal/disruption of the L2 loop including the ‐35 element (clone D), the removal of the ‐35 element (clone E), and both ‐35 and ‐10 elements (clone F). Clone G contained only the Shine–Dalgarno sequence and served as the control/comparator in promoter activity studies.
