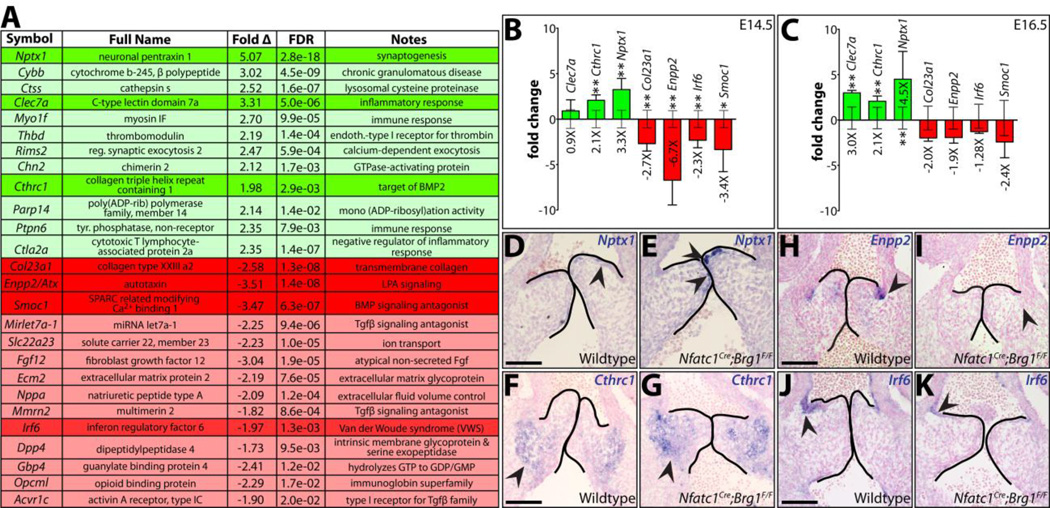
Figure 7. RNA-seq identifies novel semilunar valve expressed transcripts misexpressed in endocardial lineage Brg1 deficient valves.
(A) Select differentially expressed genes (DEGs) identified from an RNA-seq analysis comparing two paired samples of wildtype and Nfatc1Cre;Brg1F/F dissected E14.5 heart valve tissue. 26 DEGs (green-upregulated; red-downregulated) are listed with fold change and false discovery rate (FDR). Transcripts of particular interest are highlighted. (B,C) Quantitative RT-PCR expression studies of the highlighted DEGs in (A) E14.5 (B) and E16.5 (C) valves. Bar heights represent the log-base-2 fold change in gene expression (linear mean fold change value indicated below bars) between Nfatc1Cre;Brg1F/F and wildtype tissue. Error bars represent one standard deviation with the additional error bars centered along the x-axis showing the variation between wildtype samples. Double asterisks indicate a significant difference including a Bonferroni correction (one-tailed Student’s t-test, n=7, P < 0.007); single asterisk indicates a P < 0.05. (D–K) RNA in situ hybridizations for indicated transcripts on pulmonic valve sections of E13.5 wildtype or littermate Nfatc1Cre;Brg1F/F embryos. Arrowheads highlight areas of highest transcript expression. Scale bar: 100 µm.