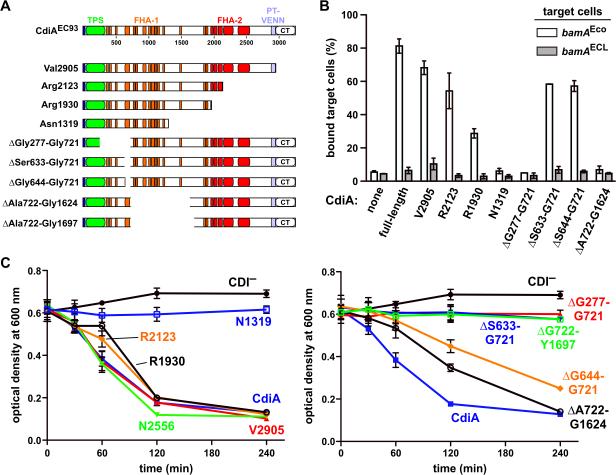
Figure 6. Receptor-dependent and -independent adhesion domains map to distinct positions within CdiAEC93.
A) Architecture of CdiAEC93. The CdiAEC93 is composed of an N-terminal TPS transport domain (green) followed by two distinct regions of filamentous hemagglutinin peptide repeats (FHA-1 and FHA-2). The pretoxin-VENN (PT-VENN) domain demarcates the C-terminal toxin region. The various CdiAEC93 truncations and in-frame deletions are shown schematically. B) BamAEco-dependent target-cell binding assays. DL4259 cells that carry pTNC-WEB (none) or the indicated CDI cosmids were mixed at a 5:1 ratio with CH9604 (bamAEco) and CH9591 (bamAECL) target cells that express DsRed. The suspensions were analyzed by flow cytometry using FL1 (533/30nm, GFP) and FL2 (585/40nm, DsRed) fluorophore filters. The percentage of bound target cells was calculated as the number of dual green/red fluorescent events divided by the total number of red fluorescent events. The average ± SEM is presented for two independent cell-binding experiments. C) Auto-aggregation of CdiAEC93 mutants. Mono-cultures of E. coli CH9591 (bamAECL) that express the indicated CdiAEC93 truncation and in-frame deletion variants were suspended and allowed to sediment at 1 × g to monitor auto-aggregation.