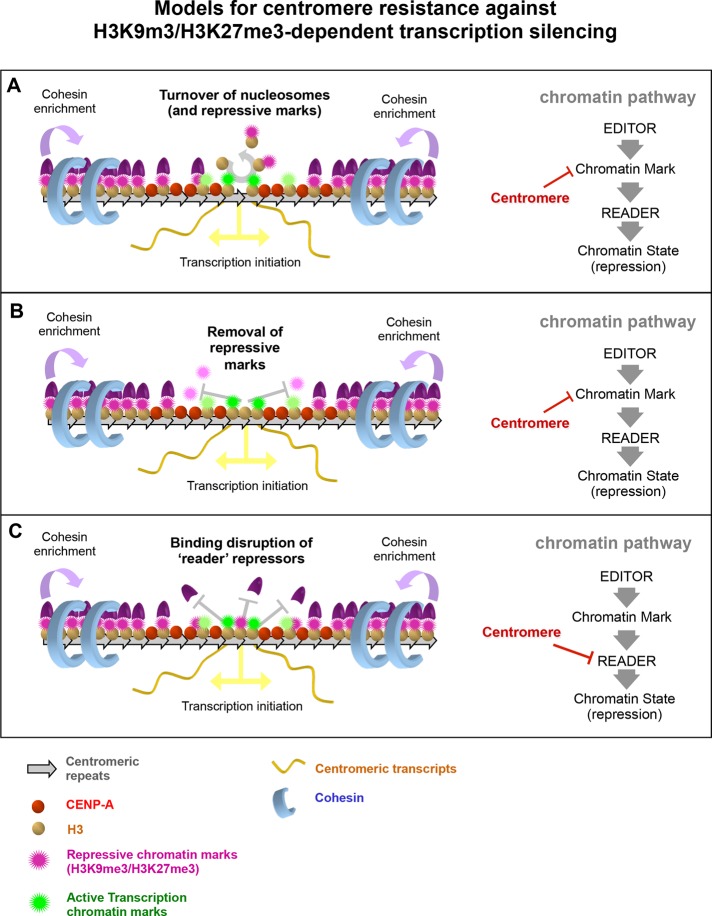
FIGURE 8:
Models for centromere resistance against H3K9m3/H3K27me3-dependent transcription silencing. The ability of eukaryote centromeres to resist local transcription silencing potentially induced by repressive chromatin marks such as H3K9me3 or H3K27me3 might may be derived from one or more properties. Our proposals for how this might occur and where in a repressive pathway the centromere may be interfering. (A) Turnover of nucleosomes within the centromere core may evict repressive chromatin marks as a consequence, thus decreasing their local concentration and potentially preventing spreading of the repressive state. (B) Direct removal of repressive chromatin marks, possibly by centromere-localized demethylases, could clear the repressive chromatin state locally at the centromere core. As in A, it could help prevent spreading of the repressed state as well. (C) Modulation of the binding of readers of repressive chromatin marks, without preventing the spreading of the repressive marks into the centromere core, can potentially alleviate transcriptional silencing promoted by those repressor readers.