Figure 1. Ubiquitin-related protein degradation by proteasome.
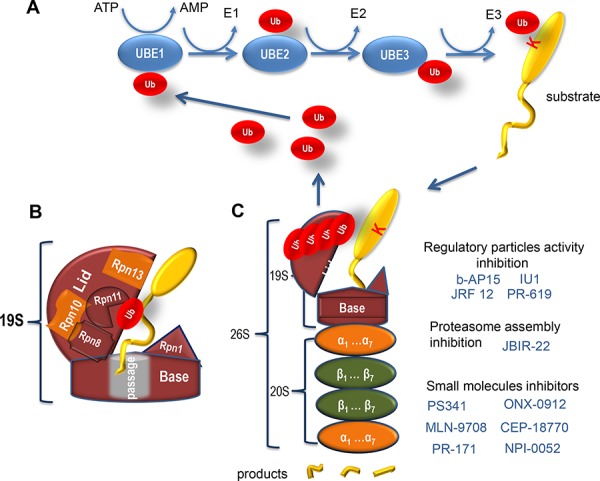
A. Shown is a scheme of the Ubiquitin Proteasome System. In general, target proteins can be covalently modified on lysine residues with one or several small (76 amino acids) proteins, called ubiquitins (Ub) (shown in red). To be transferred onto the target lysine, Ub needs to be activated first by the Ubiquitin activating enzyme (E1) by forming a thio-ester bond with the latter. This reaction requires the energy of ATP. Subsequently, Ub is transferred to one of the Ubiquitin conjugating enzymes (E2), followed by an association with a substrate-specific Ubiquitin ligase (E3) enzyme, which covalently attaches Ub to the target protein. Importantly, Ubs can modify themselves thus forming poly-Ubs chains. The target protein should be labelled by a chain of at least four Ub (poly-Ub) to be efficiently recognized by the proteasome for its subsequent degradation. B. A schematic representation of the proteasomal 19S RP. Critical subunits of the base and the lid are indicated. Rpn10 and Rnp13 ubiquitin receptor subunits are shown in orange. A substrate protein (yellow) with the ubiquitin moiety (red) is also shown. C. Shown is the schematic structure of the proteasome and small molecules (blue) that affect its different activities and the assembly. The 19S RP is shown in brown, the 20S CP comprised of alpha- and beta-type subunits (orange and green, respectively) is also presented. A substrate protein (yellow) modified with ubiquitins (red) is depicted as well as its products of degradation (yellow fragments).
