Figure 5. IRE1α RNase inhibition induces mesenchymal differentiation of glioblastoma cells and increases chemotaxis.
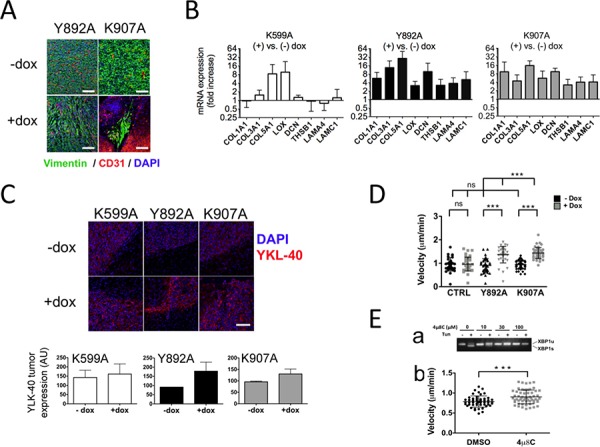
A. U87-Y892A and U87-K907A cells adopted a mesenchymal phenotype. U87-Y892A and U87-K907A-derived tumors were grown intracranially for 28 and 47 days, respectively. B. Mesenchyme-specific genes were expressed in IRE1α RNase-defective gliomas. Brain tissues (n ≥ 5 for each condition) were analyzed individually after 4 weeks of tumor development. RTqPCR analysis was carried out on a series of representative genes of the cellular matrix and of mesenchymal differentiation. Expression of the transcripts was given as fold increases in gliomas treated versus untreated with doxycycline (mean value ± SD). HPRT1 and β-actin were used as reference genes. C. Detection of YKL-40 antigen. Fluorescence intensity was quantified on two different fields of each tumor (center and rim). Three independent samples were analyzed in each group. (Bar = 100 μM.) D. Chemotaxis assays using live-cell imaging. Cells were seeded in the observation area of chemotaxis plates. One chamber was filled with DMEM/FCS/glucose, and the other with DMEM only. Individual cells were visualized in each field and chemotaxis was monitored by time-lapse for 12 h with photos taken every 15 min. Results are expressed as the average velocity (μm/sec). Ea. 4 μ8C inhibits IRE1α nuclease activity in wild-type U87 cells. Cells were exposed for 24 h to increasing amounts of 4 μ8C and presence of XBP1u and XBP1s transcripts was analyzed. Eb. 4 μ8C increased cell motility. Cells were seeded in the observation area and the two opposite chambers were filled respectively with DMEM/FBS/glucose and DMEM only. The 4 μ8C compound (20 μM) was added in both chambers. A control assay was performed in the presence of the DMSO solvent. Statistical analyzes were performed by using the Mann–Whitney test (***P < 0.001; ns, not significant).
