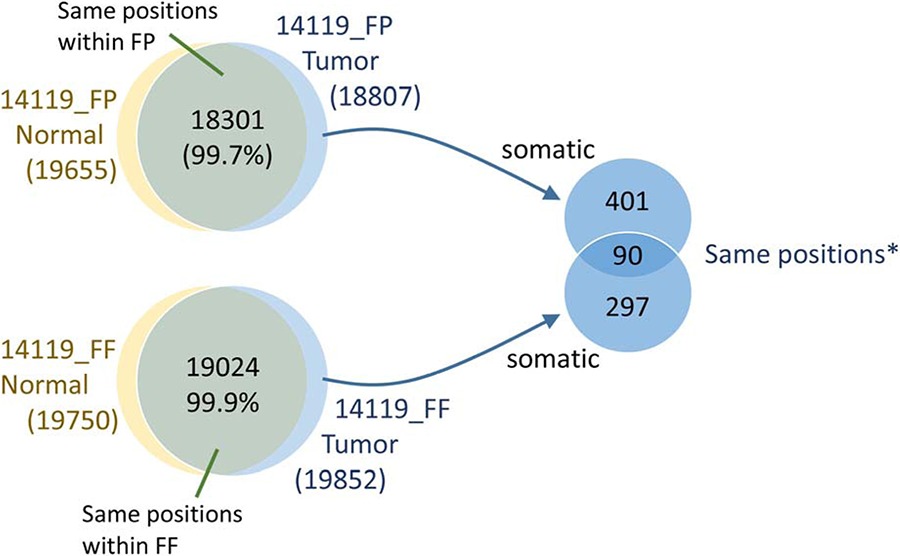
Table 1. Somatic mutations.
| WXS | Normal | Tumor | Same positions | Concordant | Somatic | Same positions* | Concordant | Discordant |
|---|---|---|---|---|---|---|---|---|
| 14119_FF | 19750 | 19852 | 19024 | 19013 | 387 | |||
| 14119_FP | 19655 | 18807 | 18301 | 18252 | 491 | 90 | 88 | 2 |
| 22285_FF | 15521 | 11253 | 10670 | 10472 | 461 | |||
| 22285_FP | 13920 | 10765 | 6296 | 6058 | 450 | 55 | 54 | 1 |
| TES | ||||||||
| 14119_FF | 1274 | 1259 | 1189 | 1188 | 30 | |||
| 14119_FP | 1266 | 1214 | 1126 | 1124 | 47 | 6 | 5 | 1 |
| 22285_FF | 2157 | 2052 | 1068 | 1063 | 42 | |||
| 22285_FP | 1219 | 881 | 416 | 408 | 49 | 5 | 5 | 0 |
SNVs in normal and tumor samples were identified in whole exome and targeted exon sequencing data sets. Overlap of positions between normal and tumor within FF or FP are indicated by “Same positions”. Overlap of positions between normal and tumor within FF and FP (all four samples) are indicated by “Same position*”. Among the overlap positions in all four samples, concordant and discordant somatic calls are shown next.