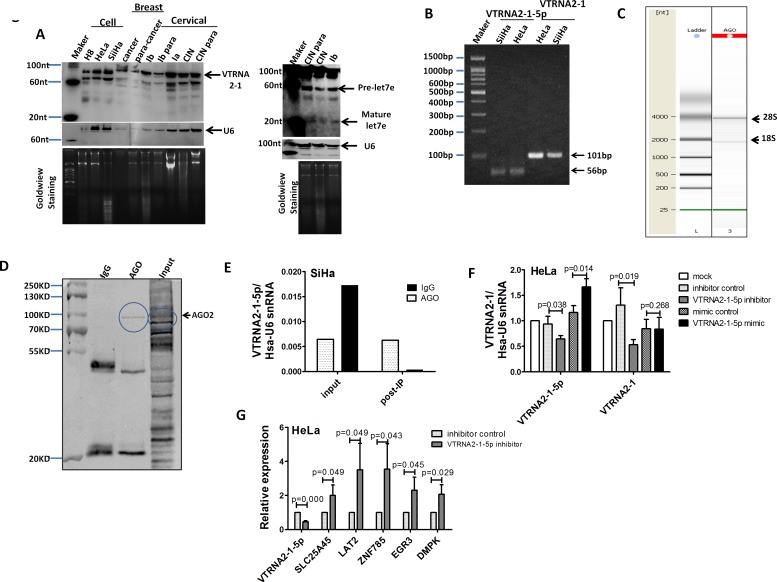
Figure 1. VTRNA2-1-5p presence in cervical tissues and cells.
A. Northern blot showing VTRNA2-1 in three cervical cell lines: one breast tumor specimen (cancer) and the surrounding normal tissue control (para-cancer); one hyperplastic cervical squamous epithelium (CINIII) and the surrounding normal tissue control (para-CINIII); and one different stage cervical cancer tissue (Ia, Ib stage) and the surrounding normal tissue control (Ib para) from the same patient. The blot was evaluated with a high-sensitivity probe specific to VTRNA2-1-5p. Human U6 was used as a loading control. Molecular size in nucleotides is indicated on the left. To rule out technical artifacts, let-7e was detected as a control in cervical tissues using a high-sensitivity probe specific to let-7e. B. After stem-loop qPCR of VTRNA2-1-5p and qRT-PCR of VTRNA2 in HeLa and SiHa, the products were resolved on a 2% agarose gel and visualized using GoldView staining. The bands consisted of the expected 102-nt band of VTRNA2 and a 54-nt band representing VTRNA2-1-5p. (C, D, E) VTRNA2-1-5p was produced through the mechanism of RISC. C. Quality check of RNAs isolated from anti-AGO-immunoprecipitated RNPs performed using a Bioanalyzer 2100 system prior to sequencing. D. Detection of AGO2 expression by Western blotting in AGO2 RNP complexes immunoprecipitated with anti-AGO2 or mouse IgG; the input samples are shown. E. Levels of VTRNA2-1-5p from RNA complex and input samples were detected with stem-loop qRT-PCR. F. Suppression of VTRNA2-1-5p in HeLa cells decreased the level of VTRNA2. G. qRT-PCR detection of mRNA expression of the predicted targets under suppression of VTRNA2-1-5p in HeLa. The results shown represent the mean±SD of at least 3 independent experiments. *p < 0.05, **p < 0.01; two-tailed Student’s t-test.