Figure 6. Effects of NKX6.3 on GKN1 expression.
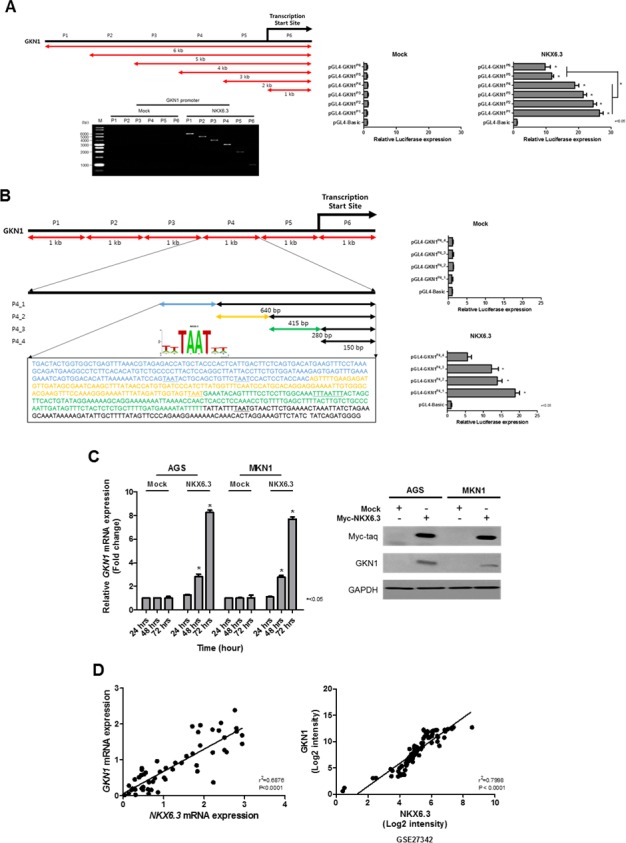
A. Putative promoter activity was characterized between −5 kb and +1 kb relative to the transcription start site (TSS) of GKN1 by ChIP and QPCR. Binding activity of NKX6.3 was detected in the GKN1 promoter region. Luciferase activity of AGSNKX6.3 cells transfected with plasmids with full-length (−5 to +1 kb) or deletions of the GKN1 promoter containing TSS designated 0 kb, then cultured for 24 h. Luciferase activity showed that NKX6.3 occupancy was 20-fold enriched at the P4 region (positions −2 to +1 kb) compared to the control. Normalized luciferase activity values for each construct (N = 3, *<0.05, t-test) are represented as mean ± SD. B. Five putative NKX6.3 binding motifs were found at the P4 promoter region. Luciferase activity analysis of 5′-deletion constructs at the P4 promoter region showed a significant decrease in promoter activity. Further deletion of the 5′ binding motifs resulted in a progressive loss of activity, indicating that NKX6.3 binding motifs at the P4 construct are required for the GKN1 transcription. C. NKX6.3 induced GKN1 mRNA and protein expression in AGSNKX6.3 and MKN1NKX6.3 cells. D. There was positive correlation between NKX6.3 and GKN1 expression in 55 non-cancerous gastric mucosa and large cohort of gastric cancer patients (NCBI GEO database, accession numbers GSE27342).
