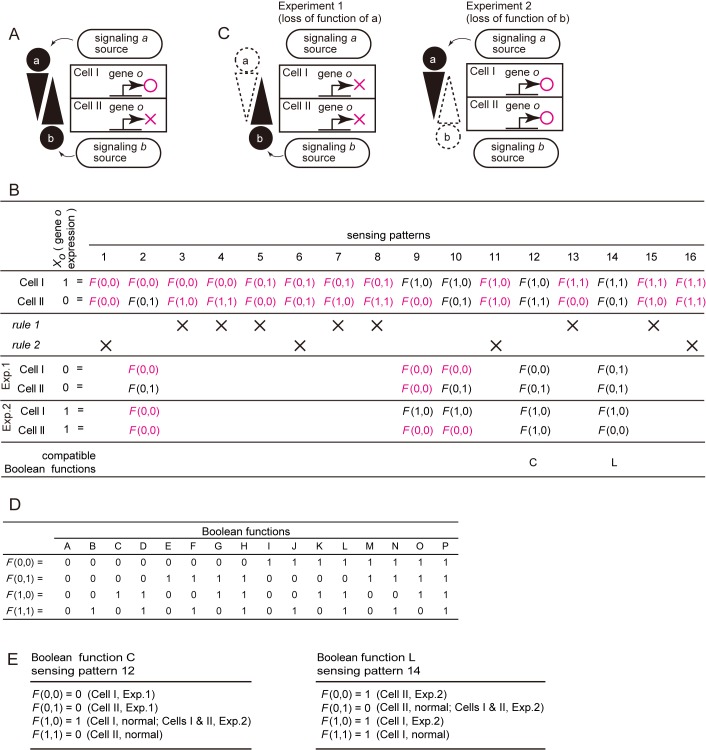
Fig 2. Boolean functions in a hypothetical biological system.
(A) A hypothetical biological system, consisting of two initially equivalent cells I and II, and two signaling molecules a and b. After a sufficient period of time, gene o is expressed only in cell I but not in cell II. (B) A Boolean function that describes expression of gene o is represented by X o = F(X a, X b), where X a and X b represent states of signaling pathways downstream of signaling molecules a and b (1, active; 0, inactive; collectively called “sensing patterns”), and X o represents expression of gene o. The 16 logically possible sensing patterns are shown in the second row. Sensing patterns and Boolean functions incompatible with Rules 1 and 2 (see text) are indicated by ‘X’ in the third row. The fourth and fifth rows show sensing patterns appearing in Experiments 1 and 2, which are shown in (C). Sensing patterns incompatible with Rules 1 and 2 are shown in magenta in the second, fourth and fifth rows. The sixth row shows a Boolean function compatible with the sensing pattern indicated in each column among all of the logically possible Boolean functions shown in (D). (C) Two conceptual loss-of-function experiments. (D) Sixteen Boolean functions that are logically possible for two input variables. (E) Two distinct combinations of Boolean functions and sensing patterns that explain the expression of gene o in this hypothetical system.