Figure 7. Post-translational modifications of Bim.
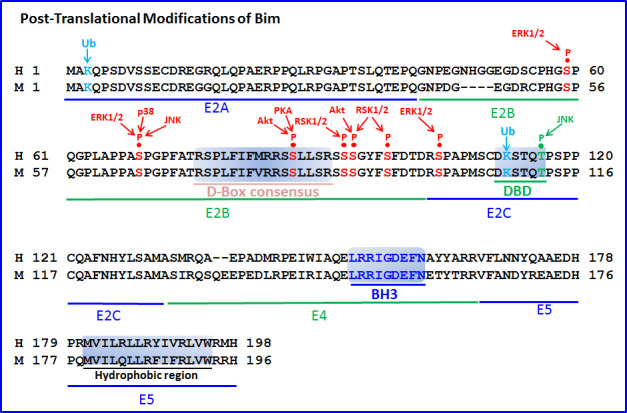
The amino acid sequences of human (H) and mouse (M) BimEL have been aligned, and important post-translational modification sites of Bim highlighted. ERK1/2 phosphorylates BimEL at three serine residues (Ser55, Ser65 and Ser100 in mouse corresponding to Ser59, Ser69 and Ser104 in human), which facilitate RSK1/2-mediated phosphorylation of Ser93/94/98 in human (corresponding to Ser89/90/94 in mouse), leading to ubiquitination at Lys3 and Lys108(M)/Lys112(H) by β-TrCP1 and proteasomal degradation. Aurora A phosphorylates the same residues as RSK1/2 during mitosis that occurs independently of previous ERK phosphorylation and leads to binding of APCCdc20 to the D-box consensus region, resulting in Bim degradation. PP2A dephosphorylates Ser93/94/98 at the exit from mitosis, thereby stabilizing BimEL. p38 and JNK also phosphorylate BimEL at Ser65 (M)/Ser69 (H), but, in addition, JNK phosphorylates Thr112(M)/Thr116(H) which lies within the dynein-binding domain (DBD). The latter phosphorylation leads to dissociation of Bim from the microtubules. JNK may also phosphorylate BimL at Thr56 (same residue as Thr112 in BimEL). p38 and JNK phosphorylation of Bim, at least in some cell types, lead to increased Bim activity. Also, Akt and PKA can phosphorylate Bim. The coding exons (E2A, E2B, E2C, E4 and E5) have been outlined, as well as the D-box consensus, DBD, BH3 domain and the hydrophobic C-terminal region.
