Figure 4. NCOA1 is recruited to the VEGFa promoter by HIF1α and c-Fos.
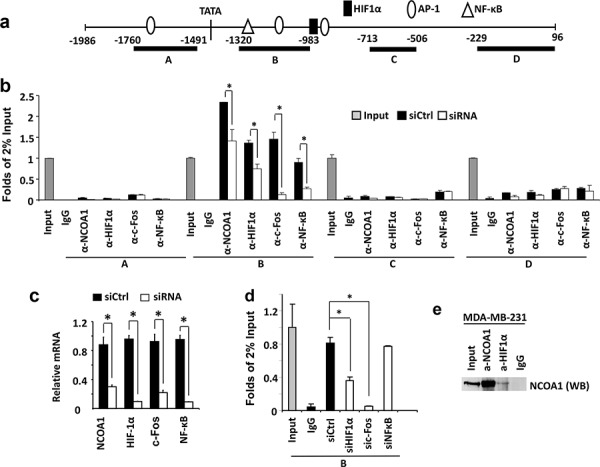
a. The VEGFa promoter region contains a TATA box and one HIF1a, three AP-1 and one NF-κB binding sites that are known to regulate VEGFa expression. Regions A–D were used for PCR amplification in ChIP assays. b. ChIP assays performed in MDA-MB-231 cells transfected with the non-targeting siRNA Smart Pool (siCtrl) or siRNA Smart Pools targeting NCOA1, HIF1α, c-Fos or NF-κB mRNAs as indicated. NCOA1, HIF1α, c-Fos and NF-κB antibodies were used for ChIP and normal IgG was used as negative control for ChIP. DNA obtained from ChIP was used as template for QPCR to measure the relative DNA amounts of regions A–D of the VEGFa promoter. The QPCR results for these regions were normalized to the QPCR results of 2% input DNA. c. The relative levels of HIF1α, c-Fos and NF-κB mRNAs in MDA-MB-231 cells transfected with non-targeting siRNA Smart Pool (siCtrl) or siRNAs targeting NCOA1, HIF1α, c-Fos and NF-κB mRNAs were measured by QPCR. d. ChIP assays for NCOA1-associated Region B of the VEGFa promoter in MDA-MB-231 cells with knockdown of HIF1α, c-Fos or NF-κB. Experiments in all panels were repeated at least three times. The * in all panels indicates p < 0.05 by Student's t test. e. Co-immunoprecipitation (Co-IP) assay for protein-protein interaction between NCOA1 and HIF1α. Cell lysate was prepared from MDA-MB-231 cells. Co-IP was performed with NCOA1 antibody, HIF1α antibody or non-immune IgG as negative control. The cell lysate for Co-IP (Input) and immunoprecipitated samples were analyzed by Western blotting (WB) using NCOA1 antibody.
