Figure 4. DNA methylation influences regulation of HIF-1α on miR-210 expression.
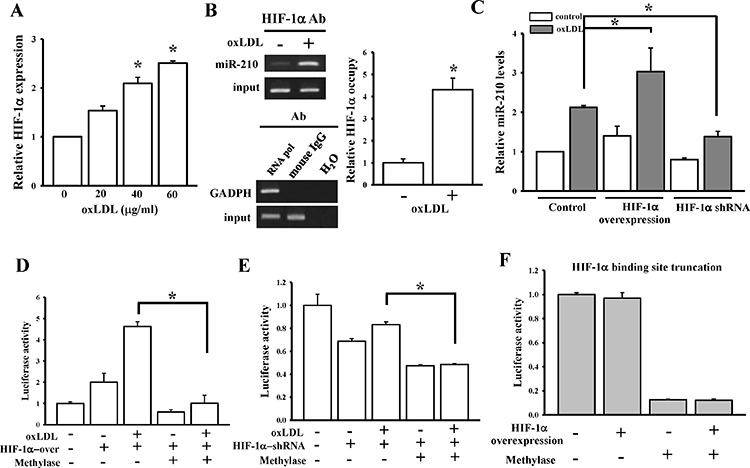
A. oxLDL effects on HIF-1α expression levels. HIF-1α expression levels were measured by a qPCR after treatment with different doses of oxLDL for 48 h. B. oxLDL effects on HIF-1α binding to the miR-210 promoter. After HASMCs were treated with 40 μg/ml oxLDL for 48 h, the HIF-1α-binding ability was measured by a ChIP assay. The right panel shows quantitative results from the ChIP assay. C. Overexpression or knockdown effects of HIF-1α on miR-210 levels. After transfection of full-length HIF-1α cDNA or HIF-1α shRNA into HASMCs and treatment with 40 μg/ml oxLDL for 48 h, the relative miR-210 expression was measured by a qPCR. The three white bars did not significantly differ. D and E. The effects of DNA methylation on HIF-1α-binding ability to the miR-210 promoter by in vitro methylation assay. HASMCs were co-transfected with a methylase-treated miR-210 promoter vector and full-length HIF-1α cDNA (or HIF-1α shRNA). After treating cells with 40 μg/ml oxLDL for 48 h, luciferase activity was measured in triplicate experiments. F. DNA methylation effects on miR-210 promoter activity with HIF-1α-binding site truncation. HASMCs were co-transfected with the methylase-treated miR-210 promoter vector in which the HIF-1α-binding site was truncated and full-length HIF-1α cDNA. After 48 h of incubation, luciferase activity was measured in triplicate experiments. Data are means ± SD of three experiments. *P < 0.05.
