Abstract
Chromosome rearrangement plays a causal role in tumorigenesis by contributing to the inactivation of tumor suppressor genes, the dysregulated expression or amplification of oncogenes and the generation of novel gene fusions. Chromosome breaks are important intermediates in this process. How, when and where these breaks arise and the specific mechanisms engaged in their repair strongly influence the resulting patterns of chromosome rearrangement. Here, we review recent progress in understanding how certain distinctive features of the cancer genome, including clustered mutagenesis, tandem segmental duplications, complex breakpoints, chromothripsis, chromoplexy and chromoanasynthesis may arise.
Genomic instability and the evolution of a cancer
Cancers evolve by natural selection. Mutations that confer increased fitness on a daughter cell contribute proportionately higher numbers to subsequent generations of cancer cells. In the context of the evolving cancer, “fitness” embraces accelerated growth, suppression of cell death, acquisition of metastatic capabilities and the other recognized hallmarks of cancer [1]. Unlike normal cells, which have evolved complex regulatory networks in support of multicellular organismal viability, the cancer cell is bound by no such constraints and is free to occupy any niche that its physiology and the host environment will allow. In this regard, the cancer cell resembles a parasitic microorganism. A certain level of genomic instability may increase the robustness of a population of microorganisms living under varying selective conditions. Similarly, genomic instability in the cancer cell, accompanied by waves of selection, may enable the cancer cell population to adapt rapidly to changing host environments during tumor growth, dissemination and metastasis [2]. Indeed, the high frequency of genomic instability in certain cancers, notably in solid tumors, suggests that this process plays a key role in the development of a mature metastatic cancer.
Recent advances in genome sequencing have revolutionized our understanding of the cancer genome and have shown that cancer-associated chromosome rearrangements are more numerous and more complex than was previously imagined [3]. Research into how these rearrangements arise is yielding exciting new insights relevant to both basic and translational arenas. First, it has revealed interesting parallels between defective double strand break (DSB) repair in cancer and model organisms. Second, it has unearthed new mechanisms connecting defective cell cycle progression with genomic instability. Third, it is expanding the universe of molecular targets and biomarkers for cancer therapy. Central to all of these areas is the formation and repair of DSBs in tumorigenesis. Here, we review these important new discoveries and discuss how they influence our understanding of cancer.
Double strand breaks: drivers of chromosome rearrangement
An unsheltered DNA end is not stable for an extended period within the cell. DSB repair mechanisms will force its interaction with other DNA molecules (Box 1). A major cause of DSBs in replicating cells is the stalling and/or collapse of the replication fork following collision with transcription complexes or at sites of abnormal DNA structure, often combined with the action of nucleases on the stalled fork [4, 5, 6]. Nuclease action can cause chromosome breakage in numerous additional ways. In normal physiology, programmed, site-specific DSBs mediate chromosome rearrangement during meiosis [7] and initiate V(D)J recombination and class switch recombination (CSR) during immunological development [8]. Other intrinsic sources of free DNA ends include the deprotection of telomeres, such as occurs during telomere attrition during aging or in cells lacking a functional shelterin complex [9]. Extrinsic causes of chromosome breakage include oxidative stress, ionizing radiation, radiomimetic chemicals and hyperosmolality [10, 11]. A host of other genotoxins can cause DSBs indirectly, via replication fork stalling or collapse [5]. Thus, DSB formation is a fairly frequent event in normal physiology. A number of elegant model systems have been employed to study chromosome translocations induced in mammalian cells in response to defined DSBs [12, 13, 14, 15, 16]. These studies suggest that spatial proximity strongly influences the likelihood that two genomically remote DNA ends will be joined.
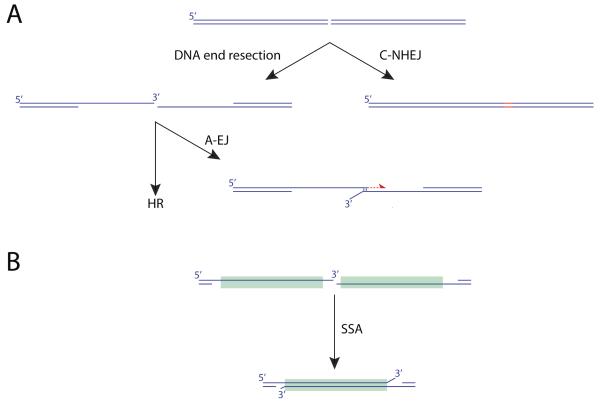
Box 1. DSB repair pathways.
The major pathways of DSB repair are classical non-homologous end joining (C-NHEJ), alternative end joining (A-EJ), single strand annealing (SSA) and homologous recombination (HR). HR, A-EJ and SSA share common initial nuclease/helicase-mediated DNA end-processing steps, generating a 3′ ssDNA intermediate, which becomes coated with the ssDNA-binding RPA heterotrimer. C-NHEJ is a rapid, high flux pathway in mammalian cells that is active throughout the cell cycle. A critical initial step is the binding of the Ku70/Ku80 heterodimer to the DNA end. A third component of this complex, the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), together with additional c-NHEJ factors, mediates synapsis between the two DNA ends. The DNA ligase IV/XRCC4 heterodimer, in conjunction with additional scaffolding proteins, ligates the DNA ends, sometimes introducing small insertions or deletions at the ligation site (shown in red). Ku binds less well to single stranded (ss)DNA tails than to blunt or minimally recessed ends. Thus, the structure of the DNA end may influence whether CNHEJ is efficiently engaged. A-EJ can rejoin two DNA ends in the absence of C-NHEJ factors. A-EJ frequently uses microhomology (MH; typically 1-5 bp) between the two DNA ends to achieve ligation. Repair synthesis (red half-arrow) can be mediated by DNA polymerase Θ. The SSA pathway mediates annealing between two ssDNA ends containing homologous direct repeats. The major features of HR are outlined in Figure 1.
Why might a cancer cell respond to DSBs differently from a normal cell? The answers are many-layered. Replication fork stalling (“replication stress”) is prevalent throughout tumorigenesis, due to the action of oncogenes, loss of cell cycle checkpoints and imbalances in cancer cell metabolism, and agents that stall replication induce copy number variations in human cells [17, 18]. Stalled replication forks and defective mitotic progression present specific challenges to the DSB repair system, as discussed in more detail below. The level of chemical damage to the cancer genome may also be elevated, for example, as a consequence of altered cancer cell metabolism. A final critical element is the configuration of the DSB repair system. Hereditary cancer syndromes provide a powerful example of the impact of DSB repair defects on cancer predisposition, by far the most prevalent examples in the human population being mutations in the hereditary breast/ovarian cancer predisposition genes BRCA1 and BRCA2. However, much of the genomic instability observed in sporadic cancers may reflect somatic inactivation of DSB repair genes, either by de novo mutation or by promoter methylation. Defective DSB repair has also emerged as a vital “Achilles’ heel” of some cancers that can be exploited for cancer therapy, as discussed below.
Pathways of DSB repair
DSB repair is commonly divided into distinct pathways of classical non-homologous end joining (C-NHEJ), alternative end joining (A-EJ), homologous recombination (HR) and single strand annealing (SSA). Box1 and Figure 1 provide an overview of these pathways in somatic cells. C-NHEJ is a high flux, rapid rejoining mechanism that results in the ligation of DNA ends without reference to the specific DNA sequence [19, 20]. A critical first step in C-NHEJ is the binding of the Ku heterodimer to the DNA ends. Ku binds avidly to double stranded (ds)DNA ends but less well to single stranded (ss)DNA tails [21]. Thus, a long ssDNA tail, if unprocessed by nucleases, may be an inefficient substrate for C-NHEJ.
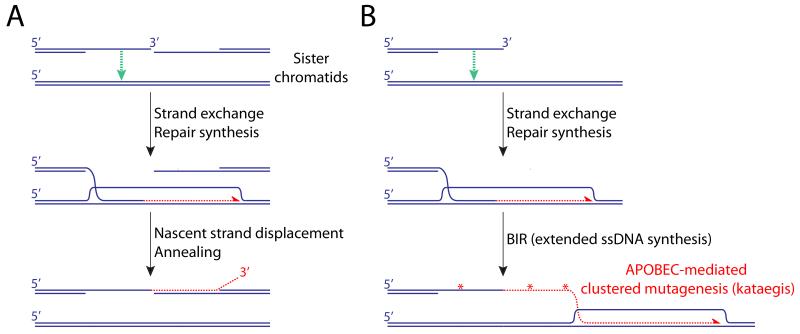
Figure 1. Homologous recombination in somatic cells.
HR mediators (primarily BRCA2 in mammalian cells and Rad52 in yeast) displace RPA from ssDNA on the resected DNA end and load Rad51, forming a nucleoprotein filament. The Rad51 filament performs a homology-seeking invasion of neighboring double stranded (ds)DNA molecules and, if a high degree of homology is detected (typically ≥100bp), a DNA polymerase extends the 3′ end of the invading (“nascent”) strand. Sequence differences copied from the donor DNA alter the sequence of the repaired DNA molecule (“gene conversion”). In S/G2 phase, the neighboring sister chromatid is the preferred donor for recombination, with potential for error-free DSB repair. However, the Rad51 filament can also detect homology at distant loci, potentially contributing to genome rearrangement. A. “Synthesis-dependent strand annealing” (SDSA) pathway of HR. Rad51-mediated strand exchange (green arrow) enables repair synthesis (red half-arrow), using the donor as template. In somatic cells, HR termination entails helicase-driven displacement of the nascent strand from the donor template followed by annealing (homologous pairing) with complementary ssDNA of the second end of the DSB. This termination mechanism does not lead to crossing over. B. Break-induced replication and kataegis. Typically triggered by a one-ended invasion, BIR is mediated by a “migrating bubble” mechanisms of leading strand synthesis (red half-arrow). The extensive tracts of ssDNA generated by BIR are potential targets of cytidine deamination (red asterisks) by APOBEC family enzymes, leading to patterns of clustered mutagenesis (kataegis).
A-EJ is a non-homologous end joining mechanism that does not require C-NHEJ genes [22, 23, 24, 25]. A-EJ-mediated rejoining is strongly biased towards the use of microhomology (MH)-mediated end joining (MMEJ), in which one or more complementary base pairs at the breakpoint are shared by the two DNA ends (Box 1) [26]. However A-EJ and MMEJ are not synonymous. For example, immune V(D)J recombination is strongly C-NHEJ-dependent but a proportion of V(D)J breakpoints in wild type cells are microhomologous. Rejoining can also be accompanied by nucleotide insertions between the two DNA ends. These insertions, which are often templated, eradicate information on breakpoint MH and have been considered to be examples of cryptic MH [27]. MH at cancer rearrangement breakpoints shows an interesting variation between different cancer types, being more prevalent in breast than prostate cancer [3, 28, 29]. The presence of MH might appear to suggest that A-EJ is the major mechanism of rejoining in cancer genome rearrangement. However, given that MH can also be a feature of C-NHEJ, additional criteria are needed to determine the mechanism. In this regard, the PolQ gene has provided tantalizing clues regarding the possible role of A-EJ in genome rearrangement.
PolQ encodes the error-prone DNA polymerase Θ. Research in model organisms has revealed a role for PolΘ in joining of DNA ends that contain extensive ssDNA tails, where rejoining is largely C-NHEJ-independent [30, 31, 32]. POLQ null mice reveal spontaneous genomic instability and fail to insert nucleotides at repair junctions during class switch recombination [30, 33]. An in vitro study showed that purified PolΘ stabilizes minimal MH by using the second end of the break as a template in trans for repair synthesis [34]. In other words, PolΘ can “broker the deal” between two DNA ends that are poor substrates for C-NHEJ and lack the extensive homology needed for homology-directed repair. Indeed, PolQ-mediated rejoining has been implicated as a mediator of chromosome rearrangement, including the fusion of dysfunctional telomeres and chromosome translocations induced by CRISPR/Cas9 breaks [35]. Cancer cells frequently reveal elevated levels of PolΘ and growth of BRCA mutant cancers is impaired by loss of PolΘ, raising the possibility that it might be a useful target for cancer therapy [35, 36].
Although deletion of key genes involved in either C-NHEJ or A-EJ promotes genomic instability [33, 37], both pathways are also implicated as mediators of pathological chromosome rearrangement [35, 38, 39]. Presumably, the availability of both C-NHEJ and A-EJ broadens the spectrum of DNA ends that can be efficiently rejoined, but these normally genome protective mechanisms can also be co-opted to mediate chromosome rearrangement in cancer.
SSA is a mechanism of joining two DNA ends that are single stranded and share extensive homology (Box 1). SSA in yeast is dependent on the HR gene RAD52 but is independent of RAD51 (the mammalian RecA homolog and central mitotic recombinase – see Figure 1), since it does not entail strand exchange [40]. The significance of SSA to cancer genome rearrangement is not well understood, since homologous breakpoints could potentially arise from either of the two “homology-directed repair” pathways of SSA or HR.
HR in somatic cells is primarily a non-crossover repair mechanism (Figure 1A). Crossing over—when it does occur—can cause loss of heterozygosity, with potential to inactivate tumor suppressor genes. During somatic HR of a two-ended DSB, mediated by the “synthesis-dependent strand annealing” (SDSA) pathway, displacement of the nascent strand and its annealing to the second DNA end normally limit gene conversion to a short tract (typically ≤100 bp) [41, 42, 43, 44]. HR also resolves “daughter strand gaps”— ssDNA gaps left in the wake of the fork [45]. When considered in the idealized case of a two-ended DSB, HR is a potentially error-free process. However, the major trigger to HR in cycling somatic cells—including cancer cells—is not an isolated two-ended DSB but a stalled or collapsed replication fork. Breakage of the stalled fork can generate a “one-ended” DSB (Figure 1B)—a DNA end that lacks a second end for the completion of DSB repair. In HR-defective cancers, this may be compounded by specific instability of the stalled replication fork. BRCA2, together with Rad51, BRCA1 and Fanconi anemia proteins protect newly synthesized DNA strands at the stalled fork from degradation by the MRE11 nuclease [46, 47]. This suggests that an HR-defective tumor suffers a “double whammy” of increased fragility of the stressed fork, in addition to the underlying DSB repair defect. In this regard, it is notable that HR is the DSB repair pathway most frequently implicated in cancer predisposition in the human population—through loss-of-function mutations of BRCA1, BRCA2, Fanconi anemia genes or a number of other HR genes [48, 49]. Indeed, a recent study identified instability of stalled forks in human cells that are haploinsufficient for BRCA1 [50], raising the exciting possibility that stalled fork instability contributes to very early stages of BRCA1-linked tumorigenesis, prior to loss of the wild type BRCA1 allele. To understand how such a defect might contribute to genomic instability, we need to consider the hazards posed by a one-ended DSB.
Replicative responses to DNA breaks: break-induced replication
A natural solution to the problem posed by a one-ended break is to reinvade the neighboring sister chromatid at the stalled replication fork and reinitiate conventional replication. Indeed, prokaryotes such as Escherichia coli achieve exactly this via an adaptor protein called PriA, which reassembles the replisome at sites of fork collapse [51]. Indeed, since E. coli have only one specified origin of replication, this process is essential for survival in the face of fork collapse. In eukaryotes, there are multiple origins of replication dispersed across the linear chromosomes and a current view is that new origins are not normally established once S phase has been initiated, even at sites of breakage and recombination. To date, no eukaryotic homolog of PriA has been identified. Thus, eukaryotes appear to lack a simple mechanism to process one-ended breaks at collapsed forks by restarting conventional replication [52].
In yeasts, one-ended breaks can trigger “break-induced replication” (BIR), a highly error-prone HR-mediated replicative response that can generate gene conversions of >100 kb [53, 54, 55]. Mapping of gene conversion tracts of spontaneous HR in Saccharomyces cerevisiae revealed a bimodal distribution of tract lengths, with median peaks at ~6 kb and >50 kb [56]. This suggests that extensive replicative responses to DSBs are common in yeast HR during normal cell growth. BIR in S. cerevisiae is mediated by POL32—a gene encoding a non-essential subunit of DNA polymerase ∂ [57, 58] and by the Pif1 helicase [59, 60]. Importantly, BIR in S. cerevisiae does not entail formation of a bona fide replication fork [60, 61]. Direct structural analysis revealed that BIR generates long tracts of single stranded DNA through a bubble migration mechanism [60]. This process is highly error-prone, introducing mutations into the newly synthesized strand at a much higher rate than during conventional replication (Figure 1B). BIR can be established following several “long tract” gene conversions (LTGCs), punctuated by template switches between homologs of the donor chromosome [62]. In the context of breaks formed at stalled/collapsed replication forks, BIR is normally limited by encounter with the adjacent replication fork (derived from the neighboring origin of replication) or by the action of the nuclease Mus81 [63]. Thus, work in model organisms has provided a framework for understanding the deleterious consequences of a one-ended break and has begun to reveal mechanisms that limit its mutational impact.
Mammalian cells can also mount extensive replicative responses during HR— experimentally measured as “long tract” gene conversion (LTGC), a process that may be analogous to BIR in yeast [64, 65, 66, 67]. Where measured, mammalian LTGC appears thus far to be limited to ~10 kb—shorter than the typical BIR tracts observed in yeast. At a conventional DSB in mammalian cells, a proportion of “one-ended” HR invasions are terminated after ~1kb of copying [66]. Thus, classical BIR is not an obligatory outcome of a one-ended HR invasion in mammalian cells. In our work on HR triggered by a site-specific replication fork barrier, loss of BRCA1 or BRCA2 paradoxically elevated the frequency of LTGC/BIR at sites of replication fork arrest, identifying the stalled fork as a significant source of aberrant replicative HR responses in mammalian cells [68]. By analogy with the work in yeast discussed above, this raises the possibility that one-ended breaks are generated more frequently at stalled forks in BRCA mutant mammalian cells [68]. Additional evidence of BIR-type copying in mammalian HR comes from analysis of copy number variation in human genomic disorders, where a BIR model was proposed to explain the formation of inverted repeats of up to ~500 kb [69].
A potential connection between mutagenesis in cancer and mammalian BIR is suggested by the phenomenon of clustered mutation or “kataegis” (thunderstorm) in cancer [70, 71]. Described simultaneously in studies of human cancer and of yeast grown under exposure to chronic alkylating damage, kataegis entails clustered mutations caused by deamination of cytosine in TpC dinucleotides (generating C->T transitions), colocalized with sites of chromosome rearrangement. Importantly, kataegis reveals “strand coordination”, whereby a switch in polarity is observed within the DNA strand from C coordination (C->T) to G coordination (G->A). These observations suggest that kataegis is organized around extensive ssDNA tracts formed with opposite polarities at sites of chromosome breakage. Consistent with this, kataegis in cancer reflects the action of APOBEC family ssDNA cytidine deaminases [72, 73, 74]. This raises the question: what is the source of ssDNA tracts that underlie kataegis? The tract length of individual kataegic clusters (up to ~200 kb) seems much greater than would be predicted to arise from conventional DNA end resection. Genetic analysis in yeast implicated the stalled replication fork as a cause of kataegis, potentially implicating one-ended breaks and BIR as an underlying mechanism. In support of this, BIR in S. cerevisiae, which entails the production of multi-kilobase tracts of ssDNA, was recently shown to reproduce kataegis-like patterns of clustered mutagenesis [75] (Figure 1B). However, the above noted switch in the polarity of strand coordination in kataegis is difficult to explain by a simple BIR model. It may be that additional mechanisms can contribute to kataegis in cancer cells.
Microhomology-mediated template switching and complex breakpoints
Although BIR is strongly RAD51-dependent in yeast [76, 77], a RAD51-independent BIR mechanism can act on short tracts of homology [78, 79]. Work in yeast [80] and in E. coli [81] identified MH-triggered replicative mechanisms for forming tandem segmental duplications (SDs). In the yeast system, generation of all SDs was dependent on POL32, suggesting that a replicative mechanism generated the SDs. SDs with homologous breakpoints were dependent on the HR gene RAD52 while those with MH breakpoints were RAD52-independent [82]. Interestingly, SD frequencies were elevated >7-fold in a rad51Δ strain. These observations are relevant to understanding the widespread tandem SDs observed in breast and ovarian cancer genomes [3, 70, 83]. Either rejoining (sister chromatid breakage/fusion) or replicative mechanisms could explain tandem SDs observed in human cancers (Figure 2).
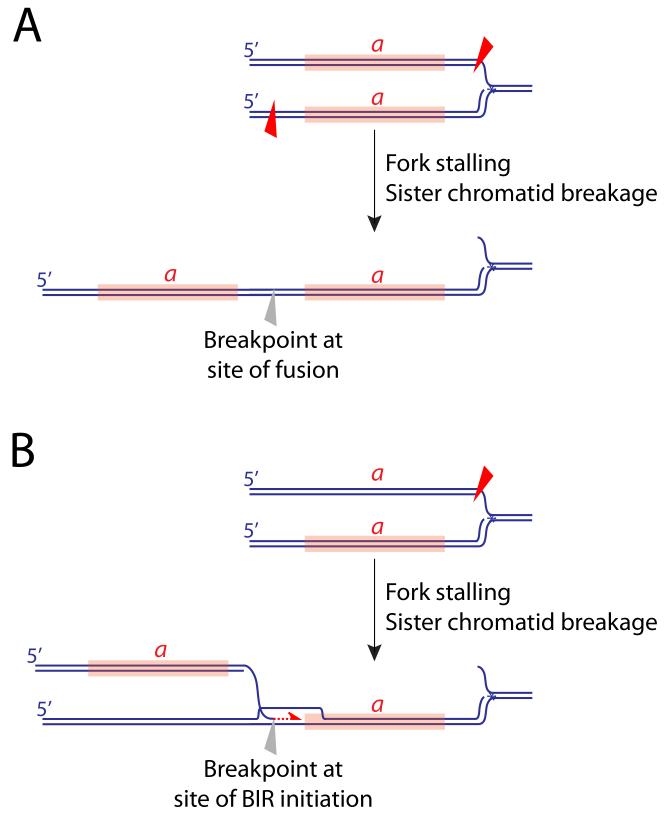
Figure 2. Mechanisms of tandem segmental duplication.
Aberrant processing of a stalled fork, perhaps augmented by an HR defect, may produce unscheduled breakage of sister chromatids in the vicinity of the stalled fork. Tandem duplication of segment a (marked in orange) could arise by asymmetrical breakage (red triangles) and fusion of sister chromatids (A), or by break-induced replication following breakage of one sister chromatid (B).
Genetic analysis in E. coli implicated MH-mediated template switching of nascent daughter strands at stalled replication forks as a trigger to SD formation [81]. MH-mediated template switching is also observed in S. cerevisiae [84]. A MH-driven template switch mechanism was proposed to explain complex breakpoints observed in the formation of PLP1 gene duplications in the human genomic disorder, Pelizaeus-Merzbacher disease [85]. The authors proposed a fork stalling and template switching model, in which a 3′ ssDNA tail undergoes template switching into the lagging strand of a spatially proximate but genomically distant replication fork [85]. This process has also been termed “microhomology-mediated break-induced replication” (MMBIR) [86], although the term “BIR” here encompasses very short tracts of copying (~hundreds of base pairs) that might be only distantly related to classical BIR as described in yeast.
These patterns of genomic instability invite direct comparison with the cancer genome. Cancer genomes frequently contain complex breakpoints at sites of rearrangement, containing arrays of short sequences derived from distinct genomic loci. Although a MH-mediated template switch mechanism (Figure 3) could explain some of these breakpoints, one study reported that complex breakpoints in cancer exhibit less MH than simple breakpoints, perhaps suggesting a simple rejoining mechanism (C-NHEJ or A-EJ) as the underlying cause [87]. However, the patterns of complex breakpoints in this study closely match those observed during A-EJ-mediated translocations induced in mammalian cells in response to two site-specific DSBs, in which a MH-mediated template switch mechanism best fits the observed data [12]. A more definitive understanding might require the development of new tools to study complex breakpoint formation in cancer.
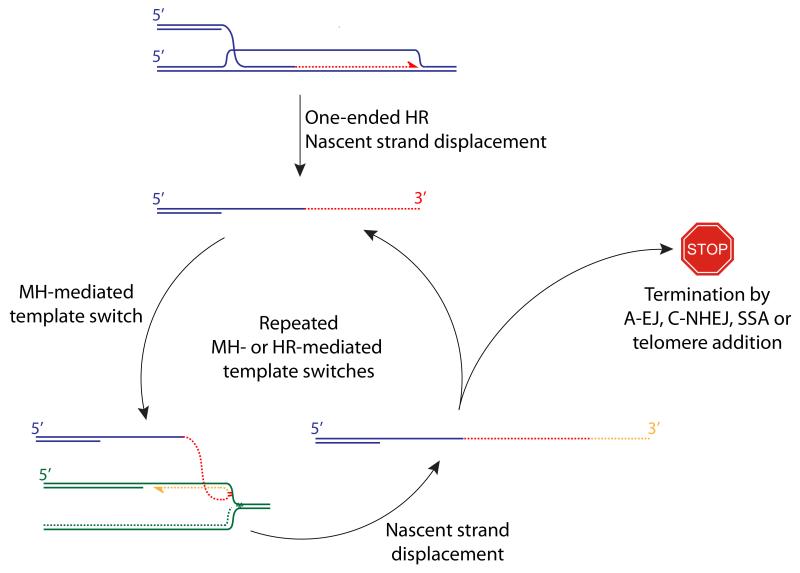
Figure 3. Origins of complex breakpoints in cancer: one-ended breaks and MH-mediated template switching.
The displaced 3′ ssDNA tail produced following a “one-ended” HR invasion may undergo MH-mediated template switching—shown here into the lagging strand of a neighboring stalled fork. Repeated rounds of MH- or HR-mediated template switches could generate the complex breakpoints observed in cancer. Termination of a template switch cycle must involve either joining to a second DNA end or telomere addition.
Connections to cancer therapy
Many effective, well established cancer chemotherapeutic agents work by stressing the replication fork. For example, the agent cisplatin forms chemical adducts on DNA, including interstrand DNA crosslinks (ICLs). ICLs present an absolute barrier to replication fork progression and HR-defective cells (BRCA mutant, Fanconi anemia mutant etc.) are hypersensitive to these agents. Thus, the largely empirical development of cancer chemotherapeutics over decades of clinical practice could be seen as an experiment in provoking intolerable levels of replication stress and lethal mitotic defects in the already overburdened cancer cell. A recent example of this is the development of inhibitors of poly(ADP-ribose) polymerase (PARP) for the treatment of HR-defective cancers. Cells lacking BRCA1, BRCA2 or other core HR genes exhibit a ~1000-fold increase in sensitivity to PARP inhibitors in comparison to wild-type cells [88, 89]. Approximately 10 years after the first demonstration of this phenomenon, PARP inhibitors were approved by the US Food and Drug Administration for the treatment of advanced BRCA-linked ovarian cancer. PARP inhibition affects multiple repair pathways and the targets that are critical for killing HR-defective cancers are not settled. However, several models invoke a mechanism that targets replication fork stability in HR-defective cells. These advances provide a strong rationale for ongoing efforts to identify the genes that normally maintain stalled fork stability, since some of these genes may be new targets for cancer therapy. A second area of translation is based on the idea that certain genomic instability “signatures” might be useful as biomarkers for cancer therapy. In this regard, the prevalence of defective HR in cancer (often termed “BRCAness”) is a particularly important phenotype. At present, there is no single biomarker of BRCAness, because of the multiplicity of genes that control HR and the frequent down-regulation of HR genes by promoter methylation rather than by mutation. Conceivably, if a specific pattern of genomic instability were to correlate with BRCA mutation status, the pattern itself might serve as a biomarker of BRCAness for cancer therapy.
Mechanisms of complex cancer genome rearrangements
Patterns of localized chromosome rearrangement in cancer cells have long been recognized as mediators of copy number variation and oncogene amplification. The classical mechanism of sequential sister chromatid breakage-fusion-bridge (BFB) cycles [90] exemplifies how complex, localized chromosome rearrangements can arise by repeated breakage and rejoining at a single locus over the course of several cell cycles, for example as a result of telomere attrition [91]. However, cancer genome rearrangements suggestive of a sudden, catastrophic event have recently been identified—termed “chromothripsis” (chromosome shattering), “chromoplexy” (braid of chromosomes) or “chromoanasynthesis” (chromosome reconstitution or chromosome reassortment) [92, 93, 94, 95, 96]. Each process might lead to sudden changes in genotype and phenotype—as exemplified by a recent cure of WHIM immunodeficiency syndrome by chromothriptic inactivation of the disease-causing dominant mutant chemokine receptor gene CXCR4 [97].
Chromothripsis was originally described in a case of chronic lymphocytic leukemia, which displayed tightly focused rearrangements on a single chromosome, with highly constrained copy-number oscillations between only two copy-number states [92, 98]. This pattern was proposed to derive from chromosome shattering, followed by the formation of tens to hundreds of locally clustered DNA rearrangements through a single event, with deletion of intervening chromosome fragments. Chromothripsis therefore is the prototype of catastrophic genome rearrangement and of punctuated genome evolution. It has since been observed in numerous cancers, often disrupting functionally important cancer genes, with gliomas showing the highest frequency of up to 39% in one study [87]. The reason for the variation in frequency of chromothripsis between different tumor types is unknown. Chromothripsis has been associated with the formation of small circular “double-minute” chromosomes, which may play a critical role in oncogene amplification [92, 99].
An intriguing insight into the mechanism underlying chromothripsis has emerged from work on micronuclei—small cytoplasmic DNA-containing bodies derived from lagging mitotic chromosomes (Figure 4A). Rupture of the micronuclear membrane could expose the micronucleated chromosome(s) to cytoplasmic endonucleases, leading to chromosome shattering [100]. Direct support for this mechanism came from single cell whole genome sequencing of two daughter cells derived from a micronucleated cell [101]. Fragments of the lagging chromosome were found reincorporated into the genomes of each daughter cell, in some cases generating complementary patterns of chromothripsis in each of the progeny cells (Figure 4A). Other rearrangements included translocations and circular derivative chromosomes—potential precursors of double minutes that carry amplified oncogenes in some cancers. Some breakpoints contained multiple short insertions (<500 bp) derived from other loci on the rearranged chromosome, suggestive of MH-mediated template switching. Work in Arabidopsis thalania has also pointed to defective mitotic progression as a cause of chromothripsis [102].
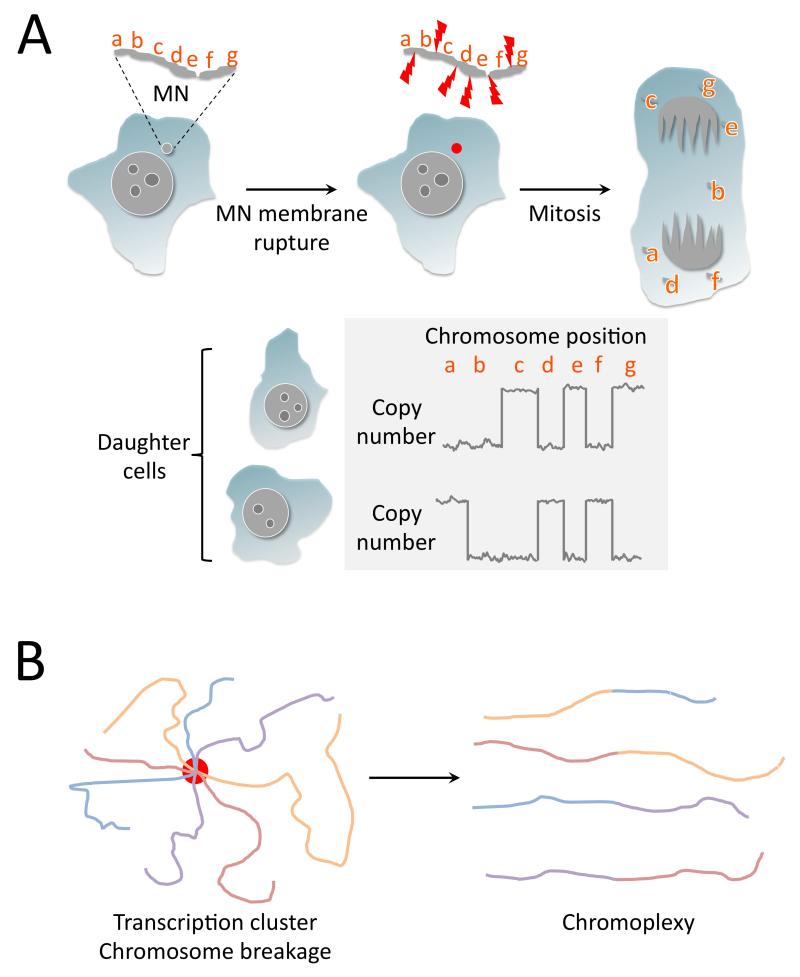
Figure 4. Chromothripsis and chromoplexy mediate catastrophic genome rearrangements via distinct mechanisms.
A. Chromothripsis can arise following mitotic damage to micronucleated chromosome(s). A micronucleated chromosome (“MN”) containing loci “a-g” is exposed to cytoplasmic nucleases when the MN membrane ruptures (red dot) and undergoes fragmentation. During the subsequent mitosis, the chromothriptic fragments segregate randomly into the two daughter cells, or are lost (e.g., fragment b shown here). The daughter cell genomes reveal constrained copy number oscillations characteristic of chromothripsis. B. A model of chromoplexy in prostate cancer. Clustering of transcriptional elements (red circle), for example, at androgen receptor (AR) responsive loci, coupled with AR-associated chromosome breakage, leads to rejoining of broken chromosoms with the production of linked translocations characteristic of chromoplexy.
Copy number neutral “constitutional chromothripsis”, generating balanced chromosome rearrangements, can cause human developmental disorders in the subsequent generation and is typically caused by de novo genome rearrangements inherited from the father [103, 104, 105]. The mechanisms of constitutional chromothripsis are currently unclear, but the copy number neutrality argues against a replication-related process. It presumably reflects extensive chromosome breakage and rejoining in the spermatid or sperm, occurring between the second meiotic division and fertilization.
Chromoplexy provides a second example of coordinated, catastrophic genome rearrangement in cancer. Like chromothripsis, chromoplexy primarily results from deletion/rejoining mechanisms. Originally described in prostate cancers, where it affects up to 40% of cases, chromoplexy entails linked rearrangements between a number of heterologous chromosomes [94]. This linkage strongly suggests that the translocations occurred in a spatially and temporally constrained fashion. The involvement of classical TMPRSS-ERG fusions at chromoplexy breakpoints suggests an underlying transcription-related mechanism. Intriguingly, androgen receptor (AR)-mediated transcription has been implicated in the formation of localized DSBs through interaction with the topoisomerase TOP2B [106]. This raises the possibility that AR transcription coordinates the induction of breaks at remote genomic loci, effectively providing the temporal and spatial linkage implied by the phenomenon of chromoplexy (Figure 4B).
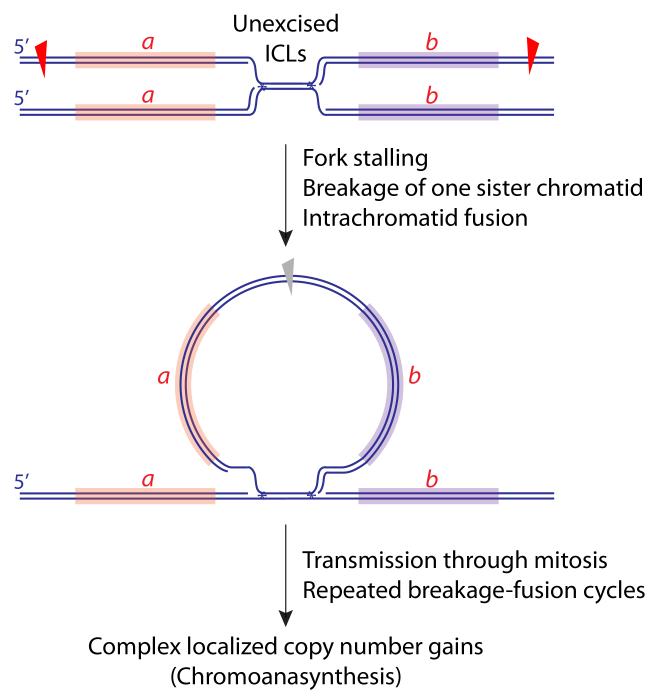
Chromoanasynthesis was described in human developmental genomic disorders as complex chromosome rearrangements with variable copy number gains at a restricted genomic locus [93]. This differs from chromothripsis and chromoplexy, which are primarily deletional/rejoining processes. Chromoanasynthesis may be part of a continuum of segmental amplification mechanisms, the tandem segmental duplication serving as the simplest element (Figure 2). A critical question is whether the copy number gains associated with chromoanasynthesis occur as a temporally discrete catastrophic event (involving BIR or aberrant re-replication of specific chromosome regions), or whether they accumulate over several cell cycles, in which case they could be explained by repeated rounds of replication with breakage-fusion cycles. The locus-specific nature of the phenomenon appears to favor the “catastrophe” model. However, an unrepaired persistent inter-strand crosslink (ICL) could cause localized copy number gains cumulatively over several cell cycles (Figure 5).
Figure 5. A model of chromoanasynthesis.
An unexcised ICL could lead to breakage of one sister chromatid, with circularization of a retained fragment, containing loci a and b, as shown. Provided the retained circular fragment lacks a centromere, it will not be a barrier to mitosis and will carry additional copies of loci a and b into the subsequent S phase. Scheduled replication, rearrangement and integration of the fragment into the gemome could contribute to the localized copy number gains characteristic of chromoanasynthesis. Notably, if the circularized fragment were to contain an origin of replication (not shown), this could further increase the opportunity for rapid copy number gains via rolling circle replication (not shown).
Intriguing insights into the mechanisms underlying chromoanasynthesis came from mutagenesis analysis in C. elegans exposed to ICL-inducing agents mechlorethamine or cisplatin [91]. In contrast to other genotoxins tested, exposure of wild type hermaphrodite parents to ICLs gave rise to offspring containing localized copy number gains. Some patterns (e.g., copy number increase from 2 to 3) could reflect simple reintegration of a retained sister chromatid fragment (Figure 5). Other patterns entailed up to 5-fold copy number increases of clustered chromosome regions 5-10 kb in size. Importantly, the timing of exposure to the genotoxins did not allow replication to occur within the hermaphrodite parent following ICL formation. Therefore, any replication-coupled ICL-induced mutagenesis must have occurred in the offspring. Since the offspring in question were not mosaics, these copy number gains likely occurred during a single S phase—the first zygotic S phase of the affected offspring. These amplifications may have arisen by repeated LTGCs of up to 10 kb at the site of the ICL. Interestingly, some examples of mammalian LTGC entail localized copy number increases with concatemer formation indicative of multiple rounds of copying of the recombining segment [65]. Perhaps intermediates of the type shown in Figure 5 could serve as templates for rolling circle replication in chromoanasynthesis.
Concluding remarks
The identification of sudden crises of chromosome rearrangement implies a potential for rapid evolution of organisms and of cancer. These processes could facilitate rapid adaptation within a species or in a population of cancer cells exposed to sudden new selective pressures. In model organisms, the capacity to undergo such rearrangements can be stress-induced [81] and similar stress-inducibility may apply to genomic catastrophes in cancer. Aberrant mitosis is increasingly recognized as a prominent cause of genomic catastrophes. Given the known relationships between replication stress—a near universal feature of cancer cells—and disordered mitotic progression, replication stress may be an underlying driving force behind both replication-associated and mitosis-associated chromosome rearrangement. The application of next generation sequencing to appropriate model systems promises to reveal these mechanisms in greater detail. As outlined in the Outstanding Questions Box, we expect that this rapidly developing field will reveal new biomarkers and new therapeutic targets in cancer.
Outstanding questions.
• Are there as yet undiscovered new types of catastrophic genome rearrangement in cancer?
• What are the genetic and mechanistic underpinnings of specific cancer genome rearrangements?
• What explains the cancer type-specific character of some chromosome rearrangements?
• Do any types of genome rearrangement in cancer correlate with tumor genotype and have potential as biomarkers in cancer? Is there a specific genome rearrangement that could serve as a biomarker of “BRCAness” across a range of different cancers?
• To what extent do rejoining vs. replicative DSB repair mechanisms contribute to cancer genome rearrangement? Do these different mechanisms vary with cancer type and do they have predictive power as biomarkers for personalized and targeted cancer therapy?
• Does the balance of DSB repair functions vary as a function of the differentiation state of the cell? Do stem cells follow established “rules” of DSB repair?
• Will the set of genes that normally suppress or promote cancer genome rearrangement reveal new targets for cancer therapy? How many new cancer therapies that exploit stalled fork instability or defective mitotic progression are awaiting discovery?
Figure I.
(associated with Box1): Diagram of major pathways involved in the repair of DNA double strand breaks (HR, C-NHEJ, A-EJ, SSA).
Advances in whole genome sequencing have provided new insight into the complexity of cancer genome rearrangements.
The discovery of catastrophic genome rearrangements in cancer establishes that genome evolution can occur in a punctuated fashion, involving temporally coordinated alterations that affect multiple distinct genetic loci.
Decoding the patterns of cancer genome rearrangement has pointed to error-prone pathways of double strand break (DSB) repair and defective mitotic progression as critical mediators of this process.
Recent advances in understanding DSB repair control provide new clues as to how specific types of cancer genome rearrangement might have arisen.
Promising new therapeutics exploit these defective DSB repair processes in the clinic.
Acknowledgments
We thank Drs. Jim Haber, Tom Petes, David Pellman, Titia de Lange, Peter Campbell, Dmitry Gordenin, Anton Gartner, Dale Ramsden, Anna Malkova, Grzegorz Ira, Jeff Sekelsky and members of the Scully lab for helpful discussions. This work was supported by an ACS postdoctoral fellowship to NAW, a Susan G. Komen grant to ER and by NIH grants R01CA095175 and R01GM073894 to RS.
Glossary
- APOBEC
“Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like.” A family of cytidine deaminases that act on cytosine in single stranded DNA.
- Break-induced replication (BIR)
a gene conversion in which up to hundreds of kilobases are copied from the donor molecule.
- Chromoanasynthesis
A chromosome rearrangement characterized by highly localized but variable increases in copy number.
- Chromoplexy
Linked translocations observed in cancer genome, suggestive of spatially and temporally coordinated break induction on multiple different chromosomes.
- Chromosome rearrangement
A type of chromosome abnormality involving a change in the structure of the native chromosome.
- Chromothripsis
A catastrophic rearrangement, often localized to one chromosome, caused by chromosome shattering. In cancer genomes, it leads to oscillations between two copy number states along the affected chromosome.
- Clustered mutagenesis
An extreme form of nonrandom distribution of mutations in the genome.
- Complex breakpoint
Breakpoints of chromosome rearrangement involving multiple loci in the reference genome rearranged into a single contiguous region of the test genome.
- Constitutional chromothripsis
Complex chromosome rearrangements carried in the germline, typically copy number neutral.
- CRISPR/Cas9
Clustered Regularly Interspaced Short Palindromic Repeats adapted to guide the Cas9 nuclease to introduce site-specific DSBs.
- Deletion
A rearrangement characterized by loss of a segment of a chromosome.
- Double-minute chromosomes
Circular fragments of DNA up to only a few megabases in size that can mediate gene amplification in cancer.
- Gene conversion
A process by which one DNA sequence replaces a homologous sequence, usually in the context of homologous recombination.
- Inter-strand crosslink (ICL)
A covalent connection of the complementary strands of DNA that prevents DNA strand separation, blocking replication and transcription.
- Kataegis
clustered hypermutation identified in some cancer genomes, mediated by the action of APOBEC cytidine deaminases on tracts of single stranded DNA.
- Long tract gene conversion (LTGC)
A gene conversion in which up to tens of kilobases are copied from the donor molecule.
- Microhomology (MH)
a type of breakpoint in which a small number of complementary base pairs at the breakpoint are shared by the two DNA ends.
- Microhomology-mediated end joining (MMEJ)
An end joining event in which the breakpoint reveals microhomology between the two DNA ends.
- Micronucleus
a small extranuclear body that contains a chromosome that was not incorporated into one of the daughter nuclei during the previous cell division.
- Segmental duplication
Generation of a second copy of a segment of DNA, located elsewhere in the genome.
- Shelterin
A complex of telomere-associated proteins that maintains normal telomere homeostasis.
- Sister chromatid
one of the two identical copies (chromatids) formed by the replication of a single chromosome.
- Synthesis-dependent strand annealing” (SDSA)
A non-crossover mechanism of homologious recombination in somatic cells.
- Tandem segmental duplication
a segmental duplication that abuts the original DNA segment “head to toe”.
- Translocation
A chromosome rearrangement between nonhomologous chromosomes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends in cell biology. 1999;9:M57–60. [PubMed] [Google Scholar]
- 3.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stratton MR. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- 6.Seigneur M, Bidnenko V, Ehrlich SD, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 7.Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annual review of genetics. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doksani Y, de Lange T. The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb Perspect Biol. 2014;6:a016576. doi: 10.1101/cshperspect.a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kultz D, Chakravarty D. Hyperosmolality in the form of elevated NaCl but not urea causes DNA damage in murine kidney cells. Proc Natl Acad Sci U S A. 2001;98:1999–2004. doi: 10.1073/pnas.98.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenburger T. DNA repair and mutagenesis. ASM Press; 2006. [Google Scholar]
- 12.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nature structural & molecular biology. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Alt FW. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–664. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–921. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Nussenzweig MC. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147:95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 18.Arlt MF, Wilson TE, Glover TW. Replication stress and mechanisms of CNV formation. Curr Opin Genet Dev. 2012;22:204–210. doi: 10.1016/j.gde.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boboila C, Alt FW, Schwer B. Classical and alternative end- joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Advances in immunology. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 20.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balestrini A, Ristic D, Dionne I, Liu XZ, Wyman C, Wellinger RJ, Petrini JH. The Ku heterodimer and the metabolism of single-ended DNA double-strand breaks. Cell reports. 2013;3:2033–2045. doi: 10.1016/j.celrep.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 23.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Roth DB. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 24.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, de Villartay JP. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. The Journal of experimental medicine. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS genetics. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends in genetics : TIG. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu AM, McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010;38:5706–5717. doi: 10.1093/nar/gkq379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Garraway LA. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drier Y, Lawrence MS, Carter SL, Stewart C, Gabriel SB, Lander ES, Getz G. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome research. 2013;23:228–235. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan SH, Yu AM, McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS genetics. 2010;6:e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koole W, van Schendel R, Karambelas AE, van Heteren JT, Okihara KL, Tijsterman M. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- 32.Roerink SF, van Schendel R, Tijsterman M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome research. 2014;24:954–962. doi: 10.1101/gr.170431.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol. 2004;24:10381–10389. doi: 10.1128/MCB.24.23.10381-10389.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nature structural & molecular biology. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, D'Andrea AD. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc Natl Acad Sci U S A. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 39.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, La Volpe A. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 40.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and molecular biology reviews : MMBR. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sweetser DB, Hough H, Whelden JF, Arbuckle M, Nickoloff JA. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol Cell Biol. 1994;14:3863–3875. doi: 10.1128/mcb.14.6.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taghian DG, Nickoloff JA. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenneman MA, Wagener BM, Miller CA, Allen C, Nickoloff JA. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Molecular Cell. 2002;10:387–395. doi: 10.1016/s1097-2765(02)00595-6. [DOI] [PubMed] [Google Scholar]
- 45.Nagaraju G, Scully R. Minding the gap: The underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair (Amst) 2007 doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H, D'Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pathania S, Bade S, Le Guillou M, Burke K, Reed R, Bowman-Colin C, Livingston DM. BRCA1 haploinsufficiency for replication stress suppression in primary cells. Nat Commun. 2014;5:5496. doi: 10.1038/ncomms6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 52.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell Cycle. 2008;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- 55.Mizuno K, Miyabe I, Schalbetter SA, Carr AM, Murray JM. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature. 2013;493:246–249. doi: 10.1038/nature11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yim E, O'Connell KE, St Charles J, Petes TD. High-resolution mapping of two types of spontaneous mitotic gene conversion events in Saccharomyces cerevisiae. Genetics. 2014;198:181–192. doi: 10.1534/genetics.114.167395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23:291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 59.Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Ira G. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donnianni RA, Symington LS. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci U S A. 2013;110:13475–13480. doi: 10.1073/pnas.1309800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 63.Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Ira G. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science. 2015;349:742–747. doi: 10.1126/science.aaa8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. Embo J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Puget N, Knowlton M, Scully R. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amst) 2005;4:149–161. doi: 10.1016/j.dnarep.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chandramouly G, Kwok A, Huang B, Willis NA, Xie A, Scully R. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat Commun. 2013;4:2404. doi: 10.1038/ncomms3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Halazonetis TD. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Willis NA, Chandramouly G, Huang B, Kwok A, Follonier C, Deng C, Scully R. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature. 2014;510:556–559. doi: 10.1038/nature13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, Lupski JR. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nature genetics. 2011;43:1074–1081. doi: 10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Stratton MR. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Gordenin DA. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nature genetics. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Gordenin DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nature genetics. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Malkova A. Break-induced replication is a source of mutation clusters underlying kataegis. Cell reports. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol Cell Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol. 2005;25:933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol Cell Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koszul R, Caburet S, Dujon B, Fischer G. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 2004;23:234–243. doi: 10.1038/sj.emboj.7600024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS genetics. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS genetics. 2008;4:e1000175. doi: 10.1371/journal.pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ng CK, Cooke SL, Howe K, Newman S, Xian J, Temple J, Brenton JD. The role of tandem duplicator phenotype in tumour evolution in high-grade serous ovarian cancer. The Journal of pathology. 2012;226:703–712. doi: 10.1002/path.3980. [DOI] [PubMed] [Google Scholar]
- 84.Anand RP, Tsaponina O, Greenwell PW, Lee CS, Du W, Petes TD, Haber JE. Chromosome rearrangements via template switching between diverged repeated sequences. Genes Dev. 2014;28:2394–2406. doi: 10.1101/gad.250258.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 86.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS genetics. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Malhotra A, Lindberg M, Faust GG, Leibowitz ML, Clark RA, Layer RM, Hall IM. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome research. 2013;23:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 89.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 90.McClintock B. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meier B, Cooke SL, Weiss J, Bailly AP, Alexandrov LB, Marshall J, Campbell PJ. C. elegans whole-genome sequencing reveals mutational signatures related to carcinogens and DNA repair deficiency. Genome research. 2014;24:1624–1636. doi: 10.1101/gr.175547.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Campbell PJ. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Bi W. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Garraway LA. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Holland AJ, Cleveland DW. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med. 2012;18:1630–1638. doi: 10.1038/nm.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang CZ, Leibowitz ML, Pellman D. Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes Dev. 2013;27:2513–2530. doi: 10.1101/gad.229559.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McDermott DH, Gao JL, Liu Q, Siwicki M, Martens C, Jacobs P, Murphy PM. Chromothriptic cure of WHIM syndrome. Cell. 2015;160:686–699. doi: 10.1016/j.cell.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 99.Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Korbel JO. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tan EH, Henry IM, Ravi M, Bradnam KR, Mandakova T, Marimuthu MP, Chan SW. Catastrophic chromosomal restructuring during genome elimination in plants. eLife. 2015;4 doi: 10.7554/eLife.06516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Cuppen E. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Human molecular genetics. 2011;20:1916–1924. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- 104.Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, Cuppen E. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell reports. 2012;1:648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 105.Weckselblatt B, Hermetz KE, Rudd MK. Unbalanced translocations arise from diverse mutational mechanisms including chromothripsis. Genome research. 2015;25:937–947. doi: 10.1101/gr.191247.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Yegnasubramanian S. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nature genetics. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]