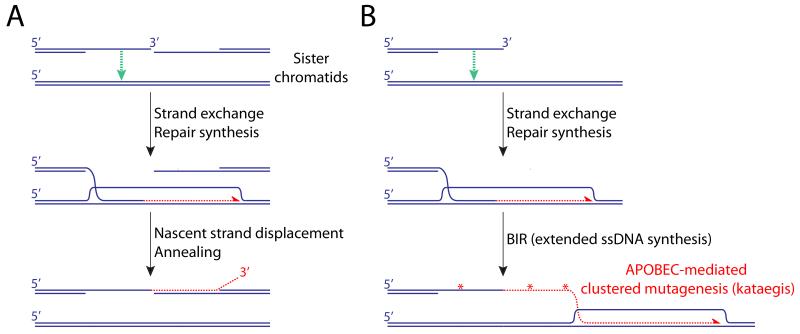
Figure 1. Homologous recombination in somatic cells.
HR mediators (primarily BRCA2 in mammalian cells and Rad52 in yeast) displace RPA from ssDNA on the resected DNA end and load Rad51, forming a nucleoprotein filament. The Rad51 filament performs a homology-seeking invasion of neighboring double stranded (ds)DNA molecules and, if a high degree of homology is detected (typically ≥100bp), a DNA polymerase extends the 3′ end of the invading (“nascent”) strand. Sequence differences copied from the donor DNA alter the sequence of the repaired DNA molecule (“gene conversion”). In S/G2 phase, the neighboring sister chromatid is the preferred donor for recombination, with potential for error-free DSB repair. However, the Rad51 filament can also detect homology at distant loci, potentially contributing to genome rearrangement. A. “Synthesis-dependent strand annealing” (SDSA) pathway of HR. Rad51-mediated strand exchange (green arrow) enables repair synthesis (red half-arrow), using the donor as template. In somatic cells, HR termination entails helicase-driven displacement of the nascent strand from the donor template followed by annealing (homologous pairing) with complementary ssDNA of the second end of the DSB. This termination mechanism does not lead to crossing over. B. Break-induced replication and kataegis. Typically triggered by a one-ended invasion, BIR is mediated by a “migrating bubble” mechanisms of leading strand synthesis (red half-arrow). The extensive tracts of ssDNA generated by BIR are potential targets of cytidine deamination (red asterisks) by APOBEC family enzymes, leading to patterns of clustered mutagenesis (kataegis).